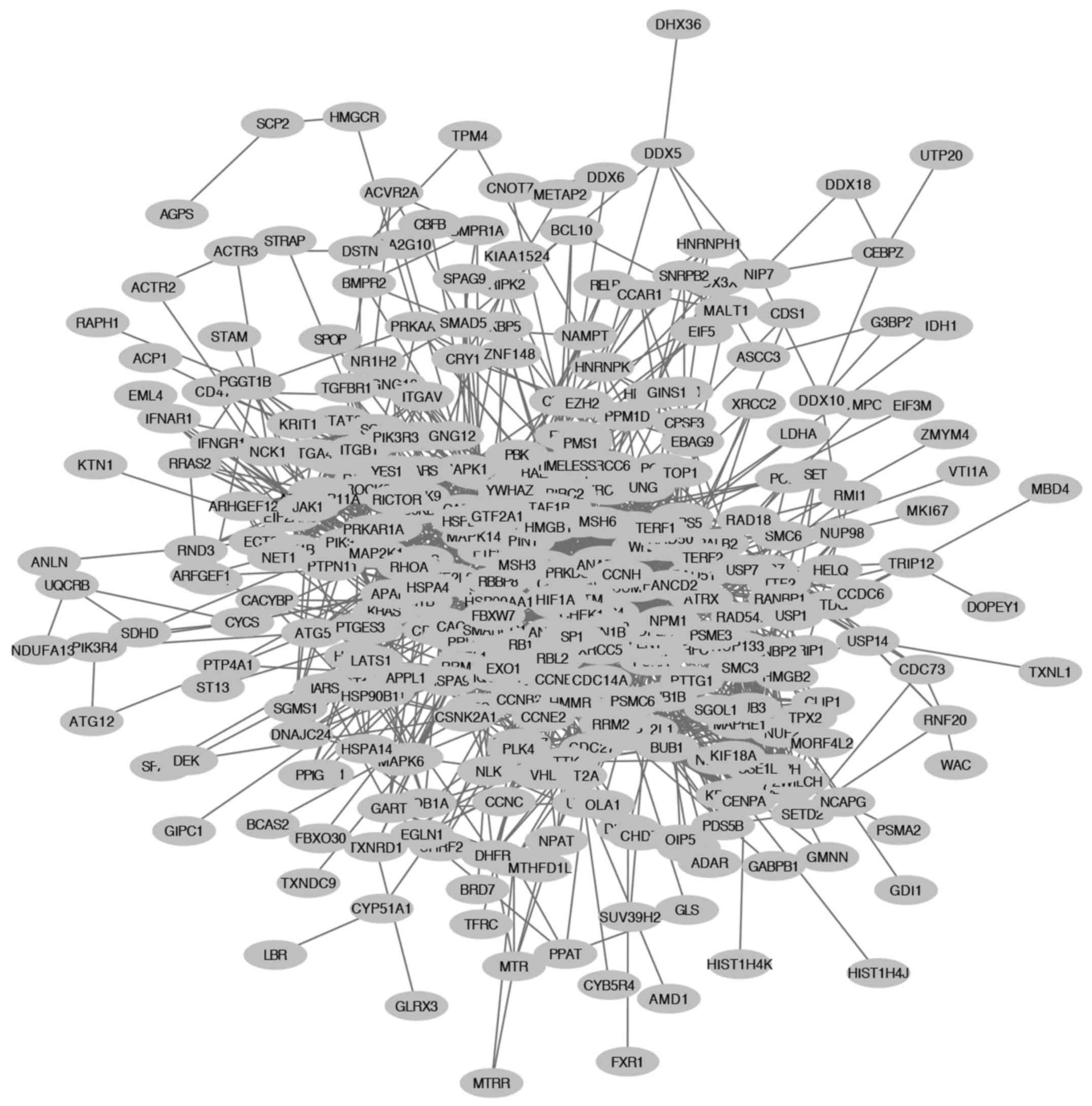
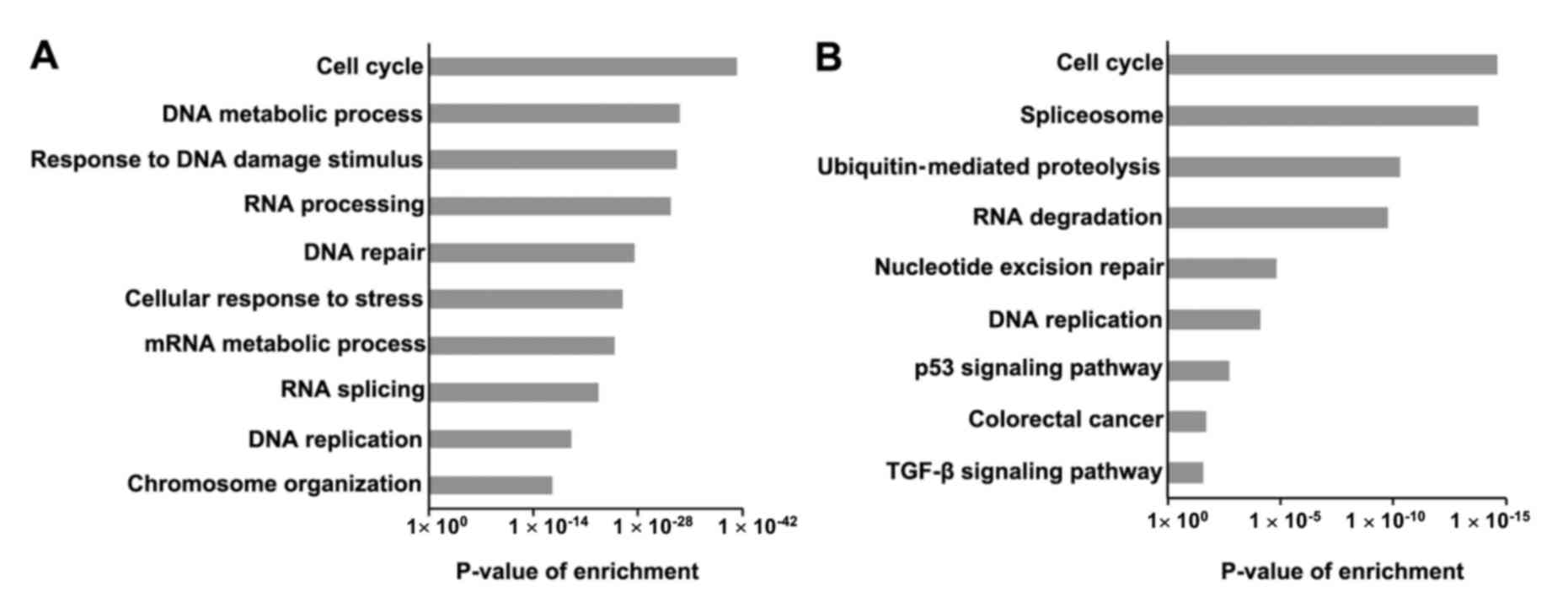
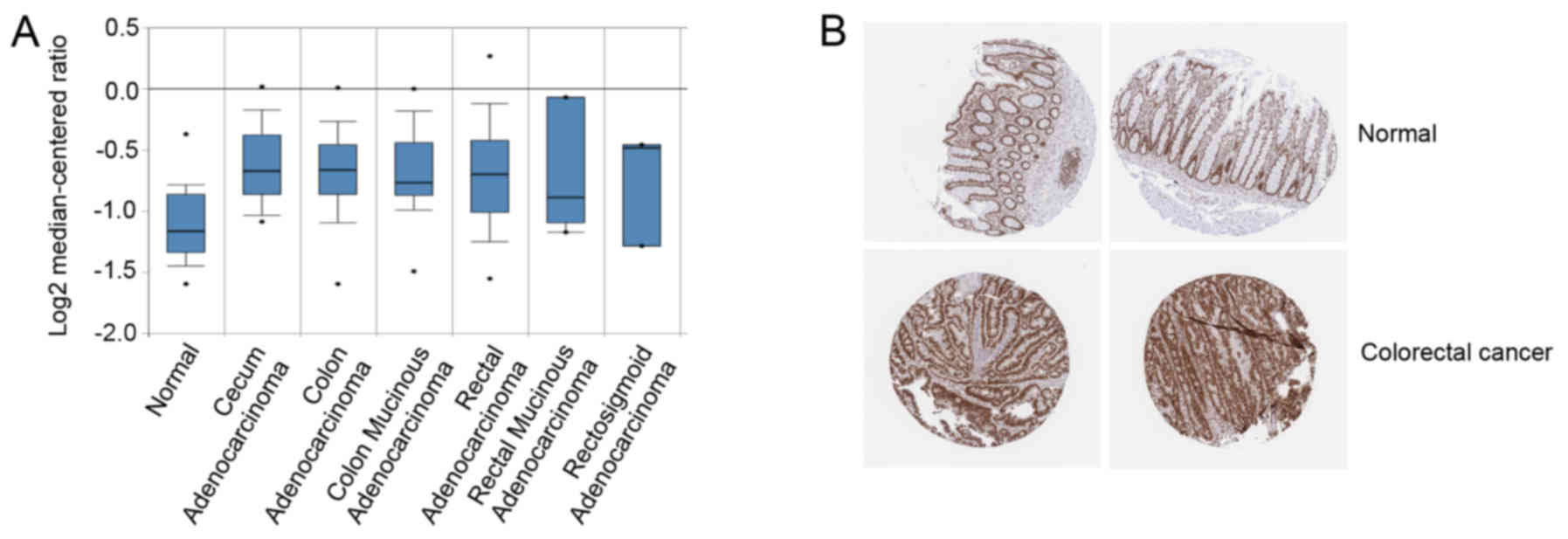
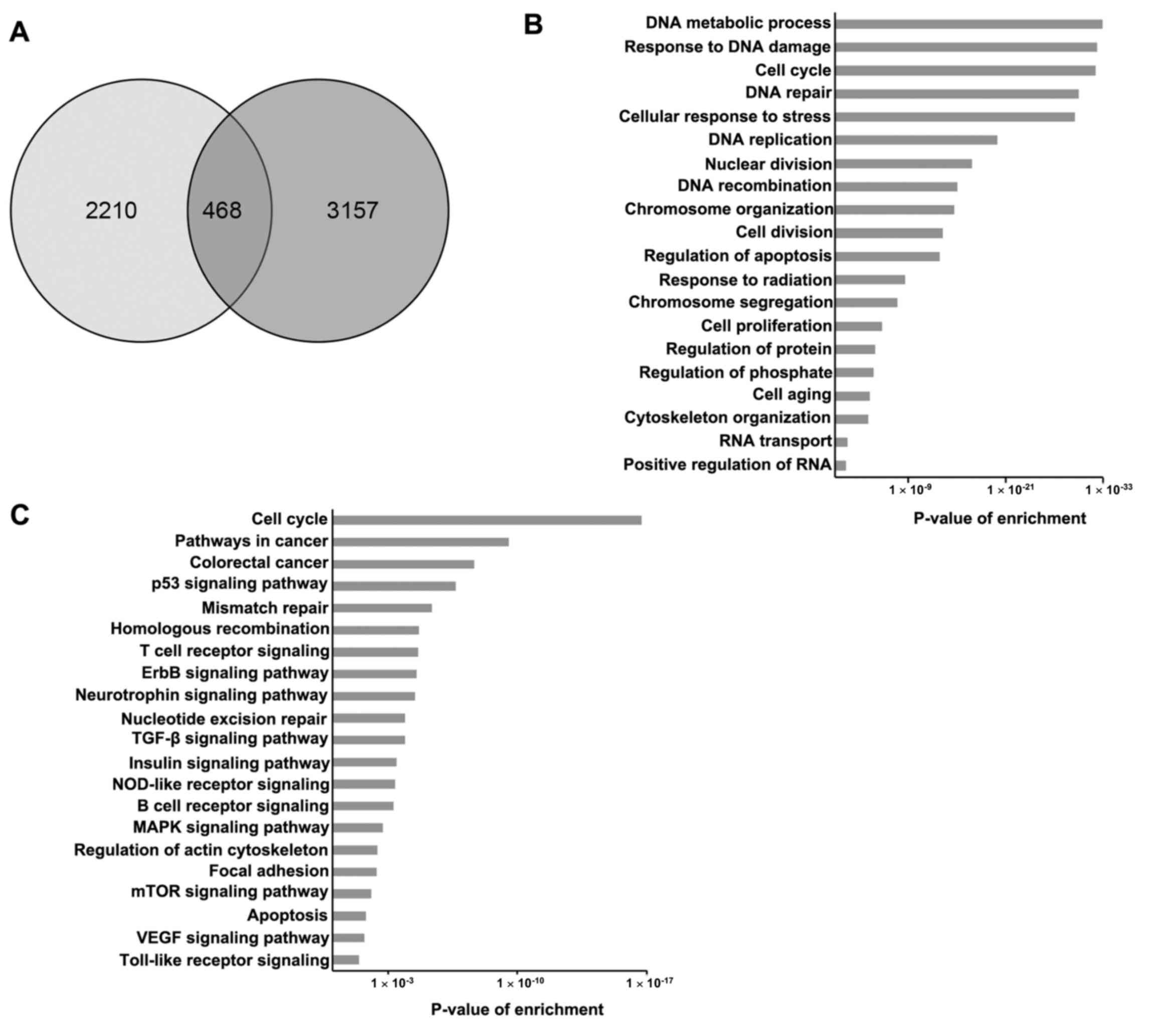
|
1
|
Sun Q, Mayeda A, Hampson RK, Krainer AR
and Rottman FM: General splicing factor SF2/ASF promotes
alternative splicing by binding to an exonic splicing enhancer.
Genes Dev. 7:2598–2608. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lemaire R, Prasad J, Kashima T, Gustafson
J, Manley JL and Lafyatis R: Stability of a PKCI-1-related mRNA is
controlled by the splicing factor ASF/SF2: A novel function for SR
proteins. Genes Dev. 16:594–607. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang Y, Gattoni R, Stevenin J and Steitz
JA: SR splicing factors serve as adapter proteins for TAP-dependent
mRNA export. Mol Cell. 11:837–843. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Z and Krainer AR: Involvement of SR
proteins in mRNA surveillance. Mol Cell. 16:597–607. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Michlewski G, Sanford JR and Cáceres JF:
The splicing factor SF2/ASF regulates translation initiation by
enhancing phosphorylation of 4E-BP1. Mol Cell. 30:179–189. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun S, Zhang Z, Sinha R, Karni R and
Krainer AR: SF2/ASF autoregulation involves multiple layers of
post-transcriptional and translational control. Nat Struct Mol
Biol. 17:306–312. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sanford JR, Gray NK, Beckmann K and
Cáceres JF: A novel role for shuttling SR proteins in mRNA
translation. Genes Dev. 18:755–768. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer
AR and Zhu J: A splicing-independent function of SF2/ASF in
microRNA processing. Mol Cell. 38:67–77. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Anczukow O, Rosenberg AZ, Akerman M, Das
S, Zhan L, Karni R, Muthuswamy SK and Krainer AR: The splicing
factor SRSF1 regulates apoptosis and proliferation to promote
mammary epithelial cell transformation. Nat Struct Mol Biol.
19:220–228. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ghigna C, Giordano S, Shen H, Benvenuto F,
Castiglioni F, Comoglio PM, Green MR, Riva S and Biamonti G: Cell
motility is controlled by SF2/ASF through alternative splicing of
the Ron protooncogene. Mol Cell. 20:881–890. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Anczuków O, Akerman M, Cléry A, Wu J, Shen
C, Shirole NH, Raimer A, Sun S, Jensen MA, Hua Y, et al:
SRSF1-regulated alternative splicing in breast cancer. Mol Cell.
60:105–117. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Karni R, de Stanchina E, Lowe SW, Sinha R,
Mu D and Krainer AR: The gene encoding the splicing factor SF2/ASF
is a proto-oncogene. Nat Struct Mol Biol. 14:185–193. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Colon Cancer Treatment (PDQ (R)): Health
Professional VersionPDQ Cancer Information Summaries. Bethesda
(MD): 2002
|
|
15
|
de Miguel FJ, Sharma RD, Pajares MJ,
Montuenga LM, Rubio A and Pio R: Identification of alternative
splicing events regulated by the oncogenic factor SRSF1 in lung
cancer. Cancer Res. 74:1105–1115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Das S, Anczukow O, Akerman M and Krainer
AR: Oncogenic splicing factor SRSF1 is a critical transcriptional
target of MYC. Cell Rep. 1:110–117. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gonçalves V, Henriques AF, Pereira JF,
Costa Neves A, Moyer MP, Moita LF, Gama-Carvalho M, Matos P and
Jordan P: Phosphorylation of SRSF1 by SRPK1 regulates alternative
splicing of tumor-related Rac1b in colorectal cells. Rna.
20:474–482. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Thorsen K, Mansilla F, Schepeler T, Øster
B, Rasmussen MH, Dyrskjøt L, Karni R, Akerman M, Krainer AR,
Laurberg S, et al: Alternative splicing of SLC39A14 in colorectal
cancer is regulated by the Wnt pathway. Mol Cell Proteomics.
10:M110.0029982011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteom. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He X, Ee PL, Coon JS and Beck WT:
Alternative splicing of the multidrug resistance protein 1/ATP
binding cassette transporter subfamily gene in ovarian cancer
creates functional splice variants and is associated with increased
expression of the splicing factors PTB and SRp20. Clin Cancer Res.
10:4652–4660. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He X, Arslan AD, Pool MD, Ho TT, Darcy KM,
Coon JS and Beck WT: Knockdown of splicing factor SRp20 causes
apoptosis in ovarian cancer cells and its expression is associated
with malignancy of epithelial ovarian cancer. Oncogene. 30:356–365.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Amin EM, Oltean S, Hua J, Gammons MV,
Hamdollah-Zadeh M, Welsh GI, Cheung MK, Ni L, Kase S, Rennel ES, et
al: WT1 mutants reveal SRPK1 to be a downstream angiogenesis target
by altering VEGF splicing. Cancer Cell. 20:768–780. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ezponda T, Pajares MJ, Agorreta J,
Echeveste JI, López-Picazo JM, Torre W, Pio R and Montuenga LM: The
oncoprotein SF2/ASF promotes non-small cell lung cancer survival by
enhancing survivin expression. Clin Cancer Res. 16:4113–4125. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Oltean S and Bates DO: Hallmarks of
alternative splicing in cancer. Oncogene. 33:5311–5318. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Silipo M, Gautrey H and Tyson-Capper A:
Deregulation of splicing factors and breast cancer development. J
Mol Cell Biol. 7:388–401. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
McCleland ML, Gardner RD, Kallio MJ, Daum
JR, Gorbsky GJ, Burke DJ and Stukenberg PT: The highly conserved
Ndc80 complex is required for kinetochore assembly, chromosome
congression and spindle checkpoint activity. Genes Dev. 17:101–114.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kobayashi Y, Takano A, Miyagi Y, Tsuchiya
E, Sonoda H, Shimizu T, Okabe H, Tani T, Fujiyama Y and Daigo Y:
Cell division cycle-associated protein 1 overexpression is
essential for the malignant potential of colorectal cancers. Int J
Oncol. 44:69–77. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hayama S, Daigo Y, Kato T, Ishikawa N,
Yamabuki T, Miyamoto M, Ito T, Tsuchiya E, Kondo S and Nakamura Y:
Activation of CDCA1-KNTC2, members of centromere protein complex,
involved in pulmonary carcinogenesis. Cancer Res. 66:10339–10348.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Numnum TM, Makhija S, Lu B, Wang M, Rivera
A, Stoff-Khalili M, Alvarez RD, Zhu ZB and Curiel DT: Improved
anti-tumor therapy based upon infectivity-enhanced adenoviral
delivery of RNA interference in ovarian carcinoma cell lines.
Gynecol Oncol. 108:34–41. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kaneko N, Miura K, Gu Z, Karasawa H,
Ohnuma S, Sasaki H, Tsukamoto N, Yokoyama S, Yamamura A, Nagase H,
et al: siRNA-mediated knockdown against CDCA1 and KNTC2, both
frequently overexpressed in colorectal and gastric cancers,
suppresses cell proliferation and induces apoptosis. Biochem
Biophys Res Commun. 390:1235–1240. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sethi G, Pathak HB, Zhang H, Zhou Y,
Einarson MB, Vathipadiekal V, Gunewardena S, Birrer MJ and Godwin
AK: An RNA interference lethality screen of the human druggable
genome to identify molecular vulnerabilities in epithelial ovarian
cancer. PLoS One. 7:e470862012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shiraishi T, Terada N, Zeng Y, Suyama T,
Luo J, Trock B, Kulkarni P and Getzenberg RH: Cancer/Testis
Antigens as potential predictors of biochemical recurrence of
prostate cancer following radical prostatectomy. J Transl Med.
9:1532011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu P, Shangguan J and Zhang L:
Downregulation of NUF2 inhibits tumor growth and induces apoptosis
by regulating lncRNA AF339813. Int J Clin Exp Pathol. 8:2638–2648.
2015.PubMed/NCBI
|
|
34
|
Sanhaji M, Friel CT, Wordeman L, Louwen F
and Yuan J: Mitotic centromere-associated kinesin (MCAK): A
potential cancer drug target. Oncotarget. 2:935–947. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ishikawa K, Kamohara Y, Tanaka F,
Haraguchi N, Mimori K, Inoue H and Mori M: Mitotic
centromere-associated kinesin is a novel marker for prognosis and
lymph node metastasis in colorectal cancer. Br J Cancer.
98:1824–1829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Di Domenico EG, Romano E, Del Porto P and
Ascenzioni F: Multifunctional role of ATM/Tel1 kinase in genome
stability: from the DNA damage response to telomere maintenance.
Biomed Res Int. 2014:7874042014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang J, Wu Q, Zhang LH, Zhao YX and Wu X:
The role of RhoA in vulvar squamous cell carcinoma: A
carcinogenesis, progression, and target therapy marker. Tumour
Biol. 37:2879–2890. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jeong D, Park S, Kim H, Kim CJ, Ahn TS,
Bae SB, Kim HJ, Kim TH, Im J, Lee MS, et al: RhoA is associated
with invasion and poor prognosis in colorectal cancer. Int J Oncol.
48:714–722. 2016. View Article : Google Scholar : PubMed/NCBI
|