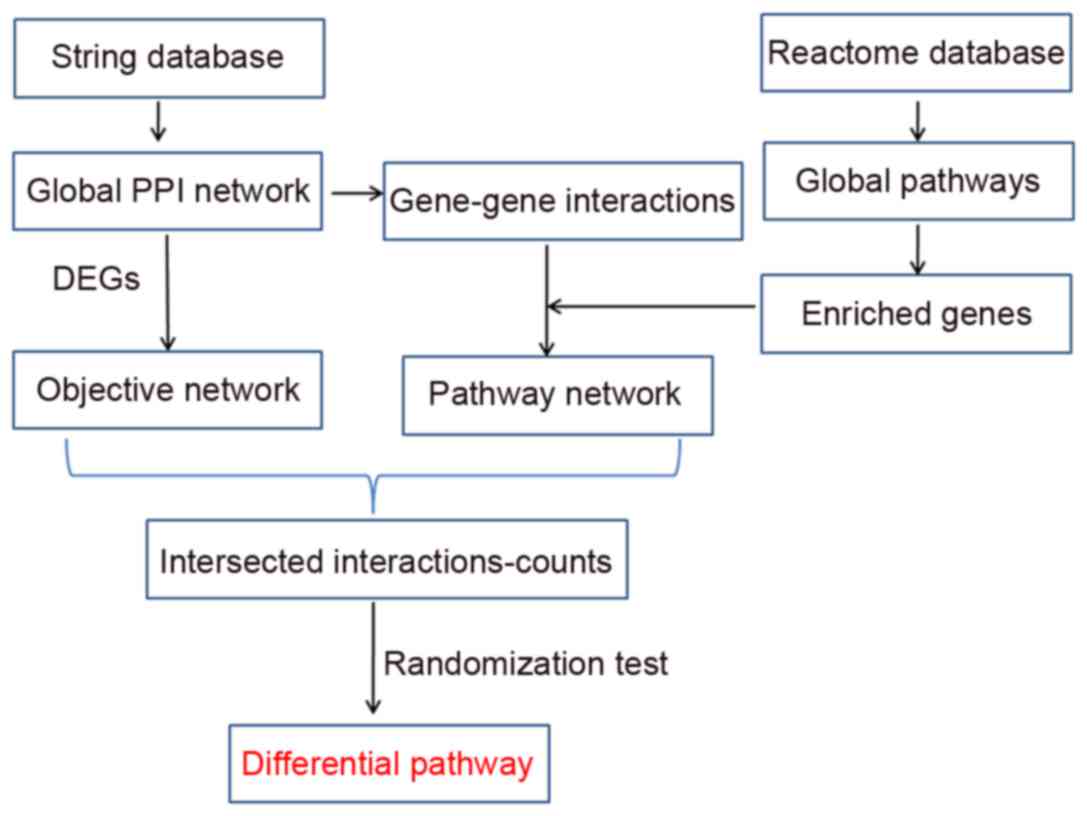
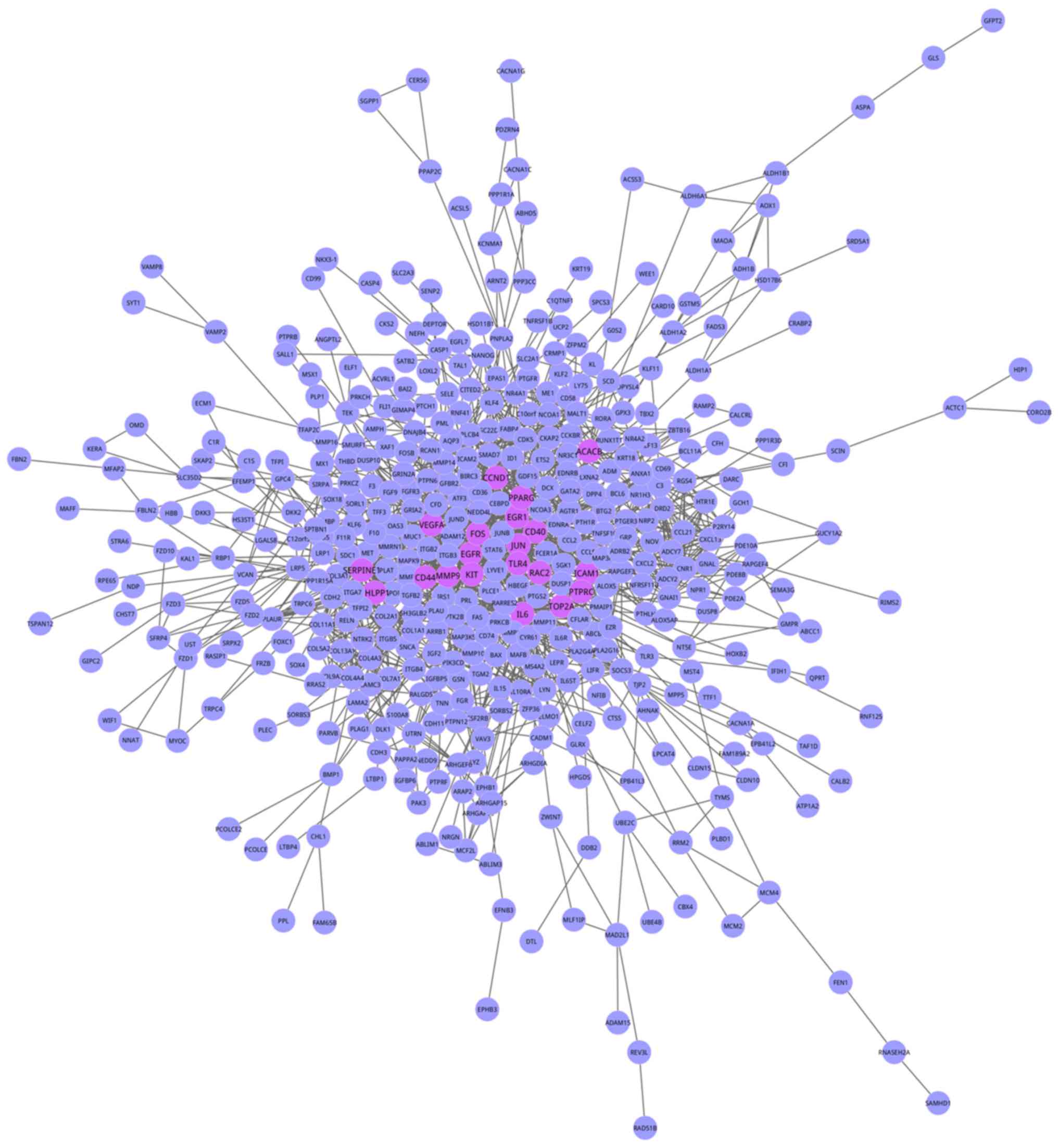
|
1
|
Islam MS, Protic O, Giannubilo SR, Toti P,
Tranquilli AL, Petraglia F, Castellucci M and Ciarmela P: Uterine
leiomyoma: Available medical treatments and new possible
therapeutic options. J Clin Endocrinol Metab. 98:921–934. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Buttram VC Jr and Reiter RC: Uterine
leiomyomata: Etiology, symptomatology, and management. Fertil
Steril. 36:433–445. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gallup DG, Blessing JA, Andersen W and
Morgan MA: Gynecologic Oncology Group Study: Evaluation of
paclitaxel in previously treated leiomyosarcoma of the uterus: A
gynecologic oncology group study. Gynecol Oncol. 89:48–51. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zivanovic O, Jacks LM, Iasonos A, Leitao
MM Jr, Soslow RA, Veras E, Chi DS, Abu-Rustum NR, Barakat RR,
Brennan MF, et al: A nomogram to predict postresection 5-year
overall survival for patients with uterine leiomyosarcoma. Cancer.
118:660–669. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jordán F, Nguyen TP and Liu WC: Studying
protein-protein interaction networks: A systems view on diseases.
Brief Funct Genomics. 11:497–504. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Koohestani F, Braundmeier AG, Mahdian A,
Seo J, Bi J and Nowak RA: Extracellular matrix collagen alters cell
proliferation and cell cycle progression of human uterine leiomyoma
smooth muscle cells. PLoS One. 8:e758442013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Halder SK, Goodwin JS and Al-Hendy A:
1,25-Dihydroxyvitamin D3 reduces TGF-beta3-induced fibrosis-related
gene expression in human uterine leiomyoma cells. J Clin Endocrinol
Metab. 96:E754–E762. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Moore AB, Yu L, Swartz CD, Zheng X, Wang
L, Castro L, Kissling GE, Walmer DK, Robboy SJ and Dixon D: Human
uterine leiomyoma-derived fibroblasts stimulate uterine leiomyoma
cell proliferation and collagen type I production, and activate
RTKs and TGF beta receptor signaling in coculture. Cell Commun
Signal. 8:102010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin CP, Chen YW, Liu WH, Chou HC, Chang
YP, Lin ST, Li JM, Jian SF, Lee YR and Chan HL: Proteomic
identification of plasma biomarkers in uterine leiomyoma. Mol
Biosyst. 8:1136–1145. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hodge JC, Kim TM, Dreyfuss JM,
Somasundaram P, Christacos NC, Rousselle M, Quade BJ, Park PJ,
Stewart EA and Morton CC: Expression profiling of uterine
leiomyomata cytogenetic subgroups reveals distinct signatures in
matched myometrium: Transcriptional profilingof the t(12;14) and
evidence in support of predisposing genetic heterogeneity. Hum Mol
Genet. 21:2312–2329. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Toyoshiba H, Yamanaka T, Sone H, Parham
FM, Walker NJ, Martinez J and Portier CJ: Gene interaction network
suggests dioxin induces a significant linkage between aryl
hydrocarbon receptor and retinoic acid receptor beta. Environ
Health Perspect. 112:1217–1224. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Toyoshiba H, Sone H, Yamanaka T, Parham
FM, Irwin RD, Boorman GA and Portier CJ: Gene interaction network
analysis suggests differences between high and low doses of
acetaminophen. Toxicol Appl Pharmacol. 215:306–316. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Barter RL, Schramm SJ, Mann GJ and Yang
YH: Network-based biomarkers enhance classical approaches to
prognostic gene expression signatures. BMC Syst Biol. 8 Suppl
4:S52014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nibbe RK, Chowdhury SA, Koyutürk M, Ewing
R and Chance MR: Protein-protein interaction networks and
subnetworks in the biology of disease. Wiley Interdiscip Rev Syst
Biol Med. 3:357–367. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu Y, Jing R, Jiang L, Jiang Y, Kuang Q,
Ye L, Yang L, Li Y and Li M: Combination use of protein-protein
interaction network topological features improves the predictive
scores of deleterious non-synonymous single-nucleotide
polymorphisms. Amino Acids. 46:2025–2035. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:(Database issue). D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Croft D, Mundo AF, Haw R, Milacic M,
Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, et al:
The Reactome pathway knowledgebase. Nucleic Acids Res. 42:(Database
issue). D472–D477. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hodge JC, Kim TM, Dreyfuss JM,
Somasundaram P, Christacos NC, Rousselle M, Quade BJ, Park PJ,
Stewart EA and Morton CC: Expression profiling of uterine
leiomyomata cytogenetic subgroups reveals distinct signatures in
matched myometrium: Transcriptional profilingof the t(12;14) and
evidence in support of predisposing genetic heterogeneity. Hum Mol
Genet. 21:2312–2329. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Barlin JN, Zhou QC, Leitao MM, Bisogna M,
Olvera N, Shih KK, Jacobsen A, Schultz N, Tap WD, Hensley ML, et
al: Molecular subtypes of uterine leiomyosarcoma and correlation
with clinical outcome. Neoplasia. 17:183–189. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of Affymetrix Genechip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bolstad B: affy: Built-in processing
methods. 2013.
|
|
23
|
Lee J and Kim DW: Efficient multivariate
feature filter using conditional mutual information. Electron Lett.
48:161–162. 2012. View Article : Google Scholar
|
|
24
|
Taminau J, Meganck S and Lazar C: Using
the inSilicoMerging package. http://www.bioconductor.org/packages//2.10/bioc/vignettes/inSilicoMerging/inst/doc/inSilicoMerging.pdfAccessed.
June 22–2012.
|
|
25
|
Smyth GK: Linear models and empirical
bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3:Article32004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhuang DY, Jiang L, He QQ, Zhou P and Yue
T: Identification of hub subnetwork based on topological features
of genes in breast cancer. Int J Mol Med. 35:664–674. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bader DA and Madduri K: Parallel
algorithms for evaluating centrality indices in real-world
networksParallel Processing. 2006, ICPP 2006, Int Confer IEEE;
Columbus, OH: pp. 539–550, 2006.
|
|
30
|
Haythornthwaite C: Social network
analysis: An approach and technique for the study of information
exchange. Lib Inf Sci Res. 18:323–342. 1996. View Article : Google Scholar
|
|
31
|
Ravasz E, Somera AL, Mongru DA, Oltvai ZN
and Barabási AL: Hierarchical organization of modularity in
metabolic networks. Science. 297:1551–1555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rifai N and Ridker PM: Proposed
cardiovascular risk assessment algorithm using high-sensitivity
C-reactive protein and lipid screening. Clin Chem. 47:28–30.
2001.PubMed/NCBI
|
|
33
|
Canay IA, Romano JP and Shaikh AM:
Randomization tests under an approximate symmetry assumption.
Econometrica. 85:1013–1030. 2017. View Article : Google Scholar
|
|
34
|
Ibragimov R and Müller UK: t-Statistic
based correlation and heterogeneity robust inference. J Business
Eco Stat. 28:453–468. 2010. View Article : Google Scholar
|
|
35
|
Jordan IK, Mariño-Ramírez L, Wolf YI and
Koonin EV: Conservation and coevolution in the scale-free human
gene coexpression network. Mol Biol Evol. 21:2058–2070. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
van Noort V, Snel B and Huynen MA: The
yeast coexpression network has a small-world, scale-free
architecture and can be explained by a simple model. EMBO Rep.
5:280–284. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Featherstone DE and Broadie K: Wrestling
with pleiotropy: Genomic and topological analysis of the yeast gene
expression network. Bioessays. 24:267–274. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Aggarwal A, Guo DL, Hoshida Y, Yuen ST,
Chu KM, So S, Boussioutas A, Chen X, Bowtell D, Aburatani H, et al:
Topological and functional discovery in a gene coexpression
meta-network of gastric cancer. Cancer Res. 66:232–241. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hynes NE, Ingham PW, Lim WA, Marshall CJ,
Massagué J and Pawson T: Signalling change: Signal transduction
through the decades. Nat Rev Mol Cell Biol. 14:393–398. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cabal-Hierro L and Lazo PS: Signal
transduction by tumor necrosis factor receptors. Cell Signal.
24:1297–1305. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Würstle ML, Laussmann MA and Rehm M: The
central role of initiator caspase-9 in apoptosis signal
transduction and the regulation of its activation and activity on
the apoptosome. Exp Cell Res. 318:1213–1220. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wynn ML, Merajver SD and Schnell S:
Unraveling the complex regulatory relationships between metabolism
and signal transduction in cancer. Adv Exp Med Biol. 736:179–189.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Makker A, Goel MM, Das V and Agarwal A:
PI3K-Akt-mTOR and MAPK signaling pathways in polycystic ovarian
syndrome, uterine leiomyomas and endometriosis: An update. Gynecol
Endocrinol. 28:175–181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Parham P: The Immune System. 4th. Garland
Science; New York, NY: 2014
|
|
45
|
Leppert P, Fouany M and Segars JH:
Understanding uterine fibroidsFibroids. Segars JH: John Wiley &
Sons, Ltd.; Oxford: 2013, View Article : Google Scholar
|
|
46
|
Wegienka G, Baird DD, Cooper T, Woodcroft
KJ and Havstad S: Cytokine patterns differ seasonally between women
with and without uterine leiomyomata. Am J Reprod Immunol.
70:327–335. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Maybin JA, Critchley HO and Jabbour HN:
Inflammatory pathways in endometrial disorders. Mol Cell
Endocrinol. 335:42–51. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Santulli P, Even M, Chouzenoux S,
Millischer AE, Borghese B, de Ziegler D, Batteux F and Chapron C:
Profibrotic interleukin-33 is correlated with uterine leiomyoma
tumour burden. Hum Reprod. 28:2126–2133. 2013. View Article : Google Scholar : PubMed/NCBI
|