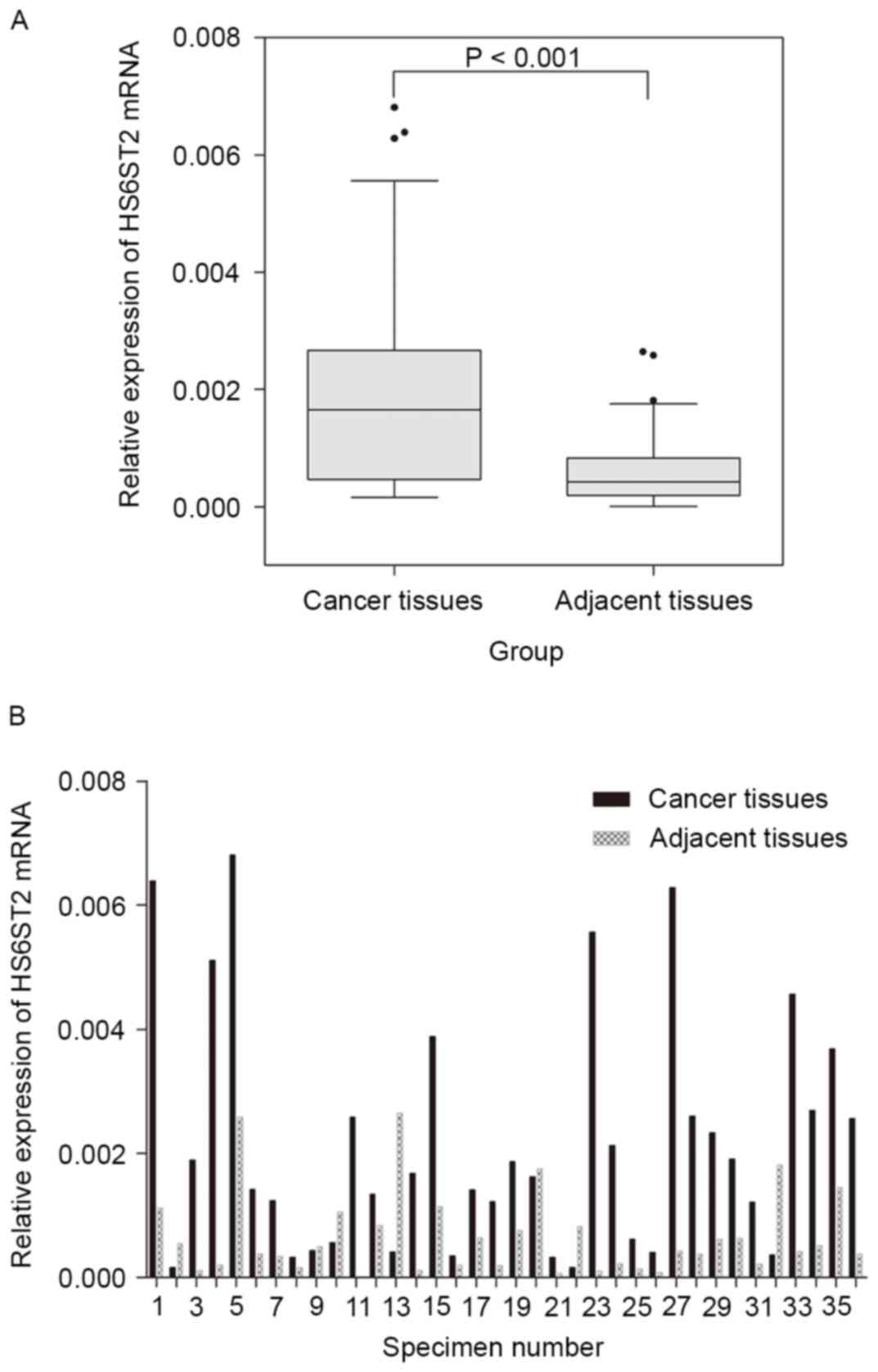
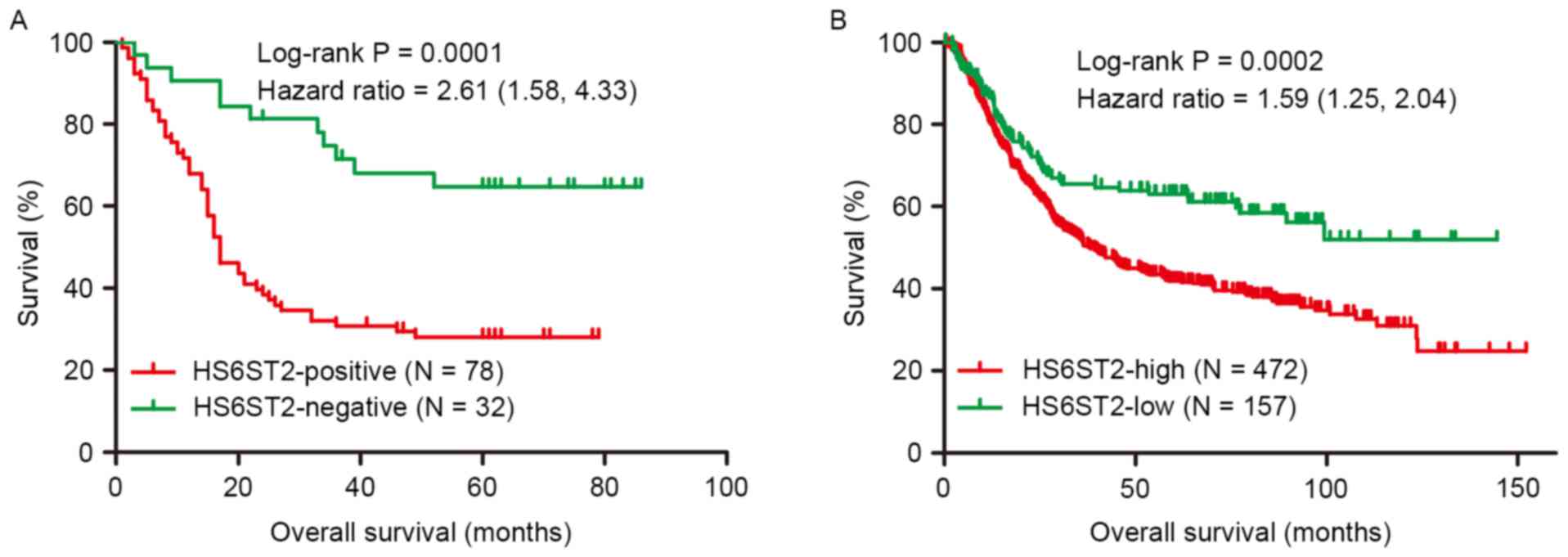
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zhang S, Zhao P, Zeng H,
Zou X and He J: Annual report on status of cancer in China, 2010.
Chin J Cancer Res. 26:48–58. 2014.PubMed/NCBI
|
|
3
|
Allemani C, Weir HK, Carreira H, Harewood
R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A,
et al: Global surveillance of cancer survival 1995–2009: Analysis
of individual data for 25,676,887 patients from 279
population-based registries in 67 countries (CONCORD-2). Lancet.
385:977–1010. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin LL, Huang HC and Juan HF: Revealing
the molecular mechanism of gastric cancer marker annexin A4 in
cancer cell proliferation using exon arrays. PLoS One.
7:e446152012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kasaian K and Jones SJ: A new frontier in
personalized cancer therapy: Mapping molecular changes. Future
Oncol. 7:873–894. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Habuchi H, Tanaka M, Habuchi O, Yoshida K,
Suzuki H, Ban K and Kimata K: The occurrence of three isoforms of
heparan sulfate 6-O-sulfotransferase having different specificities
for hexuronic acid adjacent to the targeted N-sulfoglucosamine. J
Biol Chem. 275:2859–2868. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Habuchi H, Miyake G, Nogami K, Kuroiwa A,
Matsuda Y, Kusche-Gullberg M, Habuchi O, Tanaka M and Kimata K:
Biosynthesis of heparan sulphate with diverse structures and
functions: Two alternatively spliced forms of human heparan
sulphate 6-O-sulphotransferase-2 having different expression
patterns and properties. Biochem J. 371:131–142. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nagai N, Habuchi H, Esko JD and Kimata K:
Stem domains of heparan sulfate 6-O-sulfotransferase are required
for Golgi localization, oligomer formation and enzyme activity. J
Cell Sci. 117:3331–3341. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sasisekharan R, Shriver Z, Venkataraman G
and Narayanasami U: Roles of heparan-sulphate glycosaminoglycans in
cancer. Nat Rev Cancer. 2:521–528. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Di Maro G, Orlandella FM, Bencivenga TC,
Salerno P, Ugolini C, Basolo F, Maestro R and Salvatore G:
Identification of targets of Twist1 transcription factor in thyroid
cancer cells. J Clin Endocrinol Metab. 99:E1617–E1626. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schulten HJ, Al-Mansouri Z, Baghallab I,
Bagatian N, Subhi O, Karim S, Al-Aradati H, Al-Mutawa A, Johary A,
Meccawy AA, et al: Comparison of microarray expression profiles
between follicular variant of papillary thyroid carcinomas and
follicular adenomas of the thyroid. BMC Genomics 16 Suppl.
1:S72015. View Article : Google Scholar
|
|
12
|
Hatabe S, Kimura H, Arao T, Kato H,
Hayashi H, Nagai T, Matsumoto K, DE Velasco M, Fujita Y, Yamanouchi
G, et al: Overexpression of heparan sulfate 6-sulfotransferase-2 in
colorectal cancer. Mol Clin Oncol. 1:845–850. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song K, Li Q, Peng YB, Li J, Ding K, Chen
LJ, Shao CH, Zhang LJ and Li P: Silencing of hHS6ST2 inhibits
progression of pancreatic cancer through inhibition of Notch
signalling. Biochem J. 436:271–282. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Backen AC, Cole CL, Lau SC, Clamp AR,
McVey R, Gallagher JT and Jayson GC: Heparan sulphate synthetic and
editing enzymes in ovarian cancer. Br J Cancer. 96:1544–1548. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pollari S, Käkönen RS, Mohammad KS,
Rissanen JP, Halleen JM, Wärri A, Nissinen L, Pihlavisto M,
Marjamäki A, Perälä M, et al: Heparin-like polysaccharides reduce
osteolytic bone destruction and tumor growth in a mouse model of
breast cancer bone metastasis. Mol Cancer Res. 10:597–604. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Waaijer CJ, de Andrea CE, Hamilton A, van
Oosterwijk JG, Stringer SE and Bovée JV: Cartilage tumour
progression is characterized by an increased expression of heparan
sulphate 6O-sulphation-modifying enzymes. Virchows Arch.
461:475–481. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Györffy B and Schäfer R: Meta-analysis of
gene expression profiles related to relapse-free survival in 1,079
breast cancer patients. Breast Cancer Res Treat. 118:433–441. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fekete T, Rásó E, Pete I, Tegze B, Liko I,
Munkácsy G, Sipos N, Rigó J Jr and Györffy B: Meta-analysis of gene
expression profiles associated with histological classification and
survival in 829 ovarian cancer samples. Int J Cancer. 131:95–105.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mihaly Z, Kormos M, Lánczky A, Dank M,
Budczies J, Szász MA and Győrffy B: A meta-analysis of gene
expression-based biomarkers predicting outcome after tamoxifen
treatment in breast cancer. Breast Cancer Res Treat. 140:219–232.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Orditura M, Galizia G, Sforza V,
Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J,
Savastano B, Mabilia A, et al: Treatment of gastric cancer. World J
Gastroenterol. 20:1635–1649. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Washington K: 7th edition of the AJCC
cancer staging manual: Stomach. Ann Surg Oncol. 17:3077–3079. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sawada T, Yashiro M, Sentani K, Oue N,
Yasui W, Miyazaki K, Kai K, Fushida S, Fujimura T, Ohira M, et al:
New molecular staging with G-factor supplements TNM classification
in gastric cancer: A multicenter collaborative research by the
Japan Society for Gastroenterological Carcinogenesis G-project
committee. Gastric Cancer. 18:119–128. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Harris TJ and McCormick F: The molecular
pathology of cancer. Nat Rev Clin Oncol. 7:251–265. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
D'Angelo G, Di Rienzo T and Ojetti V:
Microarray analysis in gastric cancer: A review. World J
Gastroenterol. 20:11972–11976. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bernfield M, Götte M, Park PW, Reizes O,
Fitzgerald ML, Lincecum J and Zako M: Functions of cell surface
heparan sulfate proteoglycans. Annu Rev Biochem. 68:729–777. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Robinson CJ, Harmer NJ, Goodger SJ,
Blundell TL and Gallagher JT: Cooperative dimerization of
fibroblast growth factor 1 (FGF1) upon a single heparin saccharide
may drive the formation of 2:2:1 FGF1.FGFR2c.heparin ternary
complexes. J Biol Chem. 280:42274–42282. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Robinson CJ, Mulloy B, Gallagher JT and
Stringer SE: VEGF165-binding sites within heparan sulfate encompass
two highly sulfated domains and can be liberated by K5 lyase. J
Biol Chem. 281:1731–1740. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Esko JD and Lindahl U: Molecular diversity
of heparan sulfate. J Clin Invest. 108:169–173. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cole CL, Rushton G, Jayson GC and
Avizienyte E: Ovarian cancer cell heparan sulfate
6-O-sulfotransferases regulate an angiogenic program induced by
heparin-binding epidermal growth factor (EGF)-like growth
factor/EGF receptor signaling. J Biol Chem. 289:10488–10501. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jie Zou, Peng Li, Fei Lu, Na Liu, Jianjian
Dai, Jingjing Ye, Xun Qu, Xiulian Sun, Daoxin Ma, Jino Park and
Chunyan Ji: Notch1 is required for hypoxia-induced proliferation,
invasion and chemoresistance of T-cell acute lymphoblastic leukemia
cells. J Hematol Oncol. 6:32013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang J, Mani SA, Donaher JL, Ramaswamy S,
Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and
Weinberg RA: Twist, a master regulator of morphogenesis, plays an
essential role in tumor metastasis. Cell. 117:927–939. 2004.
View Article : Google Scholar : PubMed/NCBI
|