Introduction
Renal cell carcinoma (RCC) is responsible for about
3% of all malignancies in adults, and 250,000 new cases of kidney
cancer are diagnosed each year worldwide (1). At present, clear cell RCC (ccRCC) is the
most common form of adult kidney cancer, representing a diverse set
of neoplasms with unique genetic and histological features
(2,3).
Despite developments in diagnosis and treatment strategies of RCC
during the past few years, one-third of patients present with
metastatic disease at diagnosis (2).
Furthermore, 20–40% of RCC patient that undergo surgical
nephrectomy will develop metastasis, meaning poor prognosis.
Prognostic factors for RCC include histological subtype, nuclear
grade, tumor size, local extent of the tumor and evidence of
metastatic disease at presentation (4,5). Although
a number of targeted drugs have emerged in recent years, the
overall survival times of patients with metastatic kidney cancer
remain short (6). ccRCC is generally
resistant to standard chemotherapy and radiotherapy. Previous
studies have revealed that the 5-year survival rate of patients
with metastatic RCC is <10% (7,8).
Therefore, to increase understanding of ccRCC prognosis and to
develop novel biological therapies, it is necessary to identify
molecular markers that have the potential to improve patient
outcomes and provide novel molecular targets for adjuvant
therapies.
Nudix hydroxylases (NUDTs) are a superfamily of
Mg2+-coupling enzymes found in viruses, archaea,
bacteria and eukaryotes, and catalyze the hydrolysis of nucleoside
diphosphates associated with other moieties, X (any moiety)
(9). There are two components to the
Nudix hydroxylases family: the so-called Nudix hydroxylases fold of
a β-sheet with α-helices on each side and the Nudix hydroxylases
motif which contains catalytic and metal-binding amino acids. The
Nudix hydroxylases motif is GXXXXXEXXXXXXXREUXEEXGU where U is
isoleucine, leucine or valine, and X is any amino acid (9). All NUDT family members are characterized
by a highly conserved 23-residue sequence motif, the Nudix box, and
are housecleaning enzymes (10,11). NUDT
family enzymes can activate a phosphodiester bond through the
Mg2+-assisted nucleophilic attack of a water molecule by
a basic residue. The typical NUDT reaction releases products such
as N-methyl-2-pyrrolidone, phosphate, or pyrophosphate (12,13).
The human genome has 24 NUDT hydrolase genes and
>5 pseudogenes, several of which encode more than one variant.
Expression of 17 of the 19 studied NUDT genes is strongly induced
upon entry into stationary phase, which suggests a possible
involvement in metabolic reprogramming (14,15).
Additionally, numerous site-directed mutagenesis studies have
highlighted the importance of individual residues in the Nudix
motif for catalysis. However, little is known about the NUDT family
in the field of renal cancer.
Materials and methods
NUDT expression data
Information on the expression of NUDTs and clinical
data of the Cancer Genome Atlas (TCGA) database were obtained from
the Cancer Genomics Browser of University of California Santa Cruz
(https://genome-cancer.ucsc.edu/). A
total of 24 members (NUDT1, NUDT2, NUDT3, NUDT4, NUDT5, NUDT6,
NUDT7, NUDT8, NUDT9, NUDT9P1, NUDT10, NUDT11, NUDT12, NUDT13,
NUDT14, NUDT15, NUDT16, NUDT16P1, NUDT16L1, NUDT17, NUDT18, NUDT19,
NUDT21 and NUDT22) of the NUDT family are included in the database.
In total, 509 patients (median age, 61 years; range, 26–90 years)
with primary ccRCC tumors from with detailed NUDT expression data
were chosen from the updated TCGA database according to parameters
defined in a previous study (16).
Only patients with fully characterized tumors, intact overall
survival (OS) data, complete RNAseq information and without
pretreatment were included. Clinicopathological characteristics,
including age, sex, tumor size, Tumor-Node-Metastasis (TNM) stage
(1), tumor grade, laterality,
hemoglobin level, white blood cell level, platelet level and
overall survival were collected. Follow-up of the patients was
completed with a median length of 1,063 days. In total, 347
patients succumbed during the follow-up.
Patient enrollment
For the Fudan University Shanghai Cancer Center
(FUSCC) cohort, 192 patients with ccRCC (median age, 55.5; range,
17–84 years) who underwent radical nephrectomy (RN) or nephron
sparing nephrectomy (NSS) between February 2007 and November 2011
were retrospectively enrolled. All the tissue samples were
collected during surgeries and stored at −70°C in the tissue bank
of FUSCC. The pathological subtypes were confirmed by experienced
pathologists. Clinicopathological characteristics, including age,
sex and tumor size are summarized in Table I. The present study was approved by
the Ethics Committee of Fudan University (Shanghai, China). Patient
tissues were used to investigate the expression of genes that were
thought to potentially be associated with the prognosis of patients
with ccRCC. All the patients in the present study provided signed
the informed consent for the publication of their data. All
patients provided written informed consent to their inclusion in
the study.
 | Table I.Expression of the nudix hydroxylase
family in 70 couples of paired patients in the TCGA cohort. |
Table I.
Expression of the nudix hydroxylase
family in 70 couples of paired patients in the TCGA cohort.
| Variable | TCGA cohort, n
(%) | FUSCC cohort, n
(%) |
|---|
| Total patients | 525 | 192 |
| Sex |
|
|
| Male | 341 (64.95) | 131 (68.23) |
|
Female | 184 (35.05) | 61 (31.77) |
| Grade |
|
|
| 1/2 | 240 (45.71) | 79 (41.15) |
| 3/4 | 202 (38.48) | 113 (58.85) |
| Gx | 8 (1.52) | 0 (0.00) |
| pT |
|
|
| T1 | 267 (50.86) | 129 (67.19) |
| T2 | 68 (12.95) | 29 (15.10) |
| T3 | 179 (34.10) | 27 (14.06) |
| T4 | 11 (2.10) | 7 (3.64) |
| N |
|
|
| N0 | 271 (51.62) | 181 (94.27) |
| N1 | 17 (3.24) | 4 (2.08) |
| Nx | 237 (45.14) | 7 (3.64) |
| M |
|
|
| M0 | 421 (80.19) | 184 (95.80) |
| M1 | 79 (15.05) | 7 (3.60) |
| Mx | 25 (4.76) | 1 (0.50) |
| Stage |
|
|
| I | 262 (49.90) | 130 (67.71) |
| II | 56 (10.67) | 30 (15.62) |
|
III | 126 (24.00) | 23 (11.98) |
| IV | 81 (15.43) | 9 (4.69) |
| Laterality |
|
|
|
Left | 247 (47.05) | 90 (46.87) |
|
Right | 277 (52.76) | 94 (48.95) |
|
Bilateral | 1 (0.19) | 8 (41.67) |
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
In the FUSCC cohort, total RNA was isolated from 192
ccRCC samples using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The PrimeScript RT Reagent kit
(K1622; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used
to synthesize first-strand cDNA from total RNA. Next, Synergy
Brands (SYBR)-Green real-time PCR assays (Thermo Fisher Scientific,
Inc.) were performed using an ABI 7900HT Thermocycler (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The expression level
of RNA was normalized, using relative quantification, to the level
of β-actin (17). The primers for
qPCR analysis were synthesized by Sangon Biotech Co., Ltd.
(Shanghai, China), the sequences of which are shown as follows:
NUDT5 forward, 5′-GGACTGACGCATCTGACTGT-3′ and reverse,
5′-ACAGCCGACACACACATACC-3′; NUDT9P1 forward,
5′-AGGCTGTGAACTACCGTGATG-3′ and reverse,
5′-AGAGGCTGGCATAAAGCTCA-3′; NUDT16 forward,
5′-TCTCTCCCCCAAGAAAGCATC-3′ and reverse,
5′-CCAAGGCTCACACCTCACTA-3′; NUDT17 forward,
5′-CCAACCATGGCAGAGGACAA-3′ and reverse, 5′-CTTTCCTGCTTTCCCCCGT-3′;
and β-actin forward, 5′-AGCGAGCATCCCCCAAAGTT-3′ and reverse,
5′-GGGCACGAAGGCTCATCATT-3′.
TNM stage, tumor grade, and other information were
obtained from the electronic records of the patients. Patients were
regularly followed up on the telephone or in the clinic every 3
months. Events, including tumor recurrence, progression, metastasis
and death, were recorded.
Statistical analysis
Disease-free survival was defined as time from the
date of diagnosis to the date of first recurrence or mortality. OS
was calculated from the date of diagnosis to the date of death or
of the last follow-up. Patients without events or death were
recorded as censored at the time of last follow-up. Stata 12.0
software (StataCorp LP, College Station, TX, USA) was used to
perform statistical analysis. Survival curves were constructed
using the Kaplan-Meier method, with log-rank tests used to assess
the differences between the groups. Adjusted odds ratios (ORs) with
95% confidence intervals (CIs) were calculated using the Cox
proportional hazards model. Univariate and multivariate Cox
Proportional hazards of NUDT family members expression and OS for
patients with ccRCC in the TCGA cohort were analyzed. P<0.05 was
considered to indicate a statistically significant difference.
Genes that were associated with OS were studied further.
Multivariate logistic regression was used to further study factors
that could affect the expression of NUDTs. Student's t-test or
Wilcoxon signed-rank test were performed in 70 couples of paired
patients to assess the different expression of NUDT family genes
between patients with ccRCC and healthy individuals. A t-test was
applied when the test statistic would follow a normal distribution,
if not, Wilcoxon signed-rank test was applied.
Results
Clinical characteristics of patients
with ccRCC in TCGA and FUSCC cohort
In the TCGA cohort, the median age of the 509
patients with ccRCC was 61, ranging between 26 and 90 years old. Of
these patients, 328 (64%) were male and 181 (36%) were female.
Tumor size, TNM stage, tumor grade, laterality, hemoglobin level,
white blood cell level and platelet level are shown in Table I. The median follow-up time of this
cohort was 79.5 months.
In the FUSCC cohort, the median age of these 192
patients with ccRCC was 55.5, ranging from 17 to 84 years old; 131
(68.2%) were male patients and 61 (31.8%) were female patients.
Tumor size, tumor grade, TNM stage, and tumor position are shown in
Table I. The median follow-up time of
this cohort was 47.1 months; 47 patients succumbed during
follow-up.
NUDT5, NUDT9P1, NUDT16 and NUDT17
expression were independent prognostic factors for OS in the TCGA
cohort
In univariate Cox proportion hazard ratio analysis,
age, tumor stage, metastasis, tumor stage, Fuhrman grade (All
subsequent mentions of grade are referring to Fuhrman grade),
hemoglobin level, white blood cell and platelet count, NUDT1,
NUDT3, NUDT4, NUDT5, NUDT6, NUDT7, NUDT9P1, NUDT10, NUDT11, NUDT12,
NUDT16, NUDT17, NUDT19, NUDT21 and NUDT22 expression were
significantly associated with prognosis in terms of OS of patients
with ccRCC in the TCGA cohort (P<0.05; Table II). Multivariate Cox analysis,
following adjustment for all the potential prognostic factors,
which included age, tumor stage, Fuhrman score, laterality, white
blood cell count, blood platelet count, hemoglobin content, NUDT1,
NUDT3, NUDT4, NUDT5, NUDT6, NUDT7, NUDT9P1, NUDT10, NUDT11, NUDT12,
NUDT16, NUDT17, NUDT19, NUDT21 and NUDT22, indicated that age
(HR=1.037; 95% CI, 1.020–1.053; P<0.0001), stage (HR=1.602; 95%
CI, 1.317–1.950; P<0.0001), laterality (HR=0.664; 95% CI,
0.467–0.944; P=0.023), NUDT5 (HR=1.676; 95% CI, 1.097–2.559;
P=0.017), NUDT9P1 (HR=1.512; 95% CI, 1.143–2.000; P=0.004), NUDT16
(HR=0.692; 95% CI, 0.486–0.985; P=0.041) and NUDT17 (HR=1.375; 95%
CI, 1.092–1.731; P=0.007) were the only independent predictors of
OS (all P<0.01) (Table II).
 | Table II.Univariate and multivariate Cox
Proportional Hazards analysis of integrin expression and overall
survival for patients with clear cell renal cell carcinoma in The
Cancer Genome Atlas cohort. |
Table II.
Univariate and multivariate Cox
Proportional Hazards analysis of integrin expression and overall
survival for patients with clear cell renal cell carcinoma in The
Cancer Genome Atlas cohort.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age | 1.028 | 1.015–1.042 | <0.001 | 1.036 | 1.020–1.053 | <0.001 |
| Sex | 1.073 | 0.781–1.473 | 0.665 |
|
|
|
| T | 1.964 | 1.658–2.325 | <0.001 |
|
|
|
| N | 2.799 | 1.486–5.274 | 0.001 |
|
|
|
| M | 4.448 | 3.221–6.141 | <0.001 |
|
|
|
| Stage | 1.944 | 1.695–2.229 | <0.001 | 1.603 | 1.317–1.949 | <0.001 |
| Grade | 2.350 | 1.899–2.908 | <0.001 | 1.230 | 0.920–1.644 | 0.162 |
| Hb | 0.584 | 0.415–0.823 | 0.002 | 0.915 | 0.624–1.342 | 0.651 |
| WBC | 0.652 | 0.471–0.902 | 0.010 | 1.014 | 0.694–1.483 | 0.942 |
| PLT | 1.702 | 1.145–2.529 | 0.008 | 1.086 | 0.748–1.579 | 0.664 |
| Tumor size | 1.174 | 0.946–1.459 | 0.146 |
|
|
|
| Laterality | 0.669 | 0.491–0.913 | 0.011 | 0.664 | 0.467–0.944 | 0.023 |
| NUDT1 | 1.629 | 1.346–1.971 | <0.001 | 0.942 | 0.661–1.341 | 0.740 |
| NUDT2 | 0.779 | 0.596–1.018 | 0.068 |
|
|
|
| NUDT3 | 2.089 | 1.215–3.593 | 0.008 | 0.739 | 0.401–1.361 | 0.332 |
| NUDT4 | 0.706 | 0.538–0.925 | 0.011 | 0.849 | 0.614–1.175 | 0.324 |
| NUDT5 | 2.165 | 1.684–2.783 | <0.001 | 1.676 | 1.097–2.559 | 0.017 |
| NUDT6 | 0.608 | 0.461–0.801 | <0.001 | 0.947 | 0.648–1.383 | 0.778 |
| NUDT7 | 0.716 | 0.576–0.889 | 0.003 | 1.032 | 0.730–1.457 | 0.859 |
| NUDT8 | 1.113 | 0.953–1.299 | 0.174 |
|
|
|
| NUDT9 | 0.971 | 0.639–1.474 | 0.888 |
|
|
|
| NUDT9P1 | 1.274 | 1.011–1.606 | 0.040 | 1.512 | 1.143–2.000 | 0.004 |
| NUDT10 | 1.203 | 1.074–1.347 | 0.001 | 1.180 | 0.987–1.412 | 0.069 |
| NUDT11 | 1.347 | 1.208–1.503 | <0.001 | 0.962 | 0.796–1.162 | 0.684 |
| NUDT12 | 0.661 | 0.542–0.805 | <0.001 | 0.781 | 0.600–1.017 | 0.066 |
| NUDT13 | 0.991 | 0.809–1.214 | 0.931 |
|
|
|
| NUDT14 | 0.934 | 0.778–1.121 | 0.464 |
|
|
|
| NUDT15 | 0.742 | 0.496–1.107 | 0.144 |
|
|
|
| NUDT16 | 0.677 | 0.499–0.919 | 0.012 | 0.692 | 0.486–0.985 | 0.041 |
| NUDT16P1 | 0.947 | 0.792–1.133 | 0.554 |
|
|
|
| NUDT16L1 | 1.096 | 0.813–1.478 | 0.546 |
|
|
|
| NUDT17 | 1.583 | 1.341–1.870 | <0.001 | 1.375 | 1.092–1.732 | 0.007 |
| NUDT18 | 0.975 | 0.788–1.206 | 0.817 |
|
|
|
| NUDT19 | 2.026 | 1.372–2.991 | <0.001 |
|
|
|
| NUDT21 | 0.700 | 0.519–0.946 | 0.020 | 1.113 | 0.682–1.815 | 0.668 |
| NUDT22 | 1.424 | 1.127–1.800 | 0.003 | 1.090 | 0.686–1.730 | 0.716 |
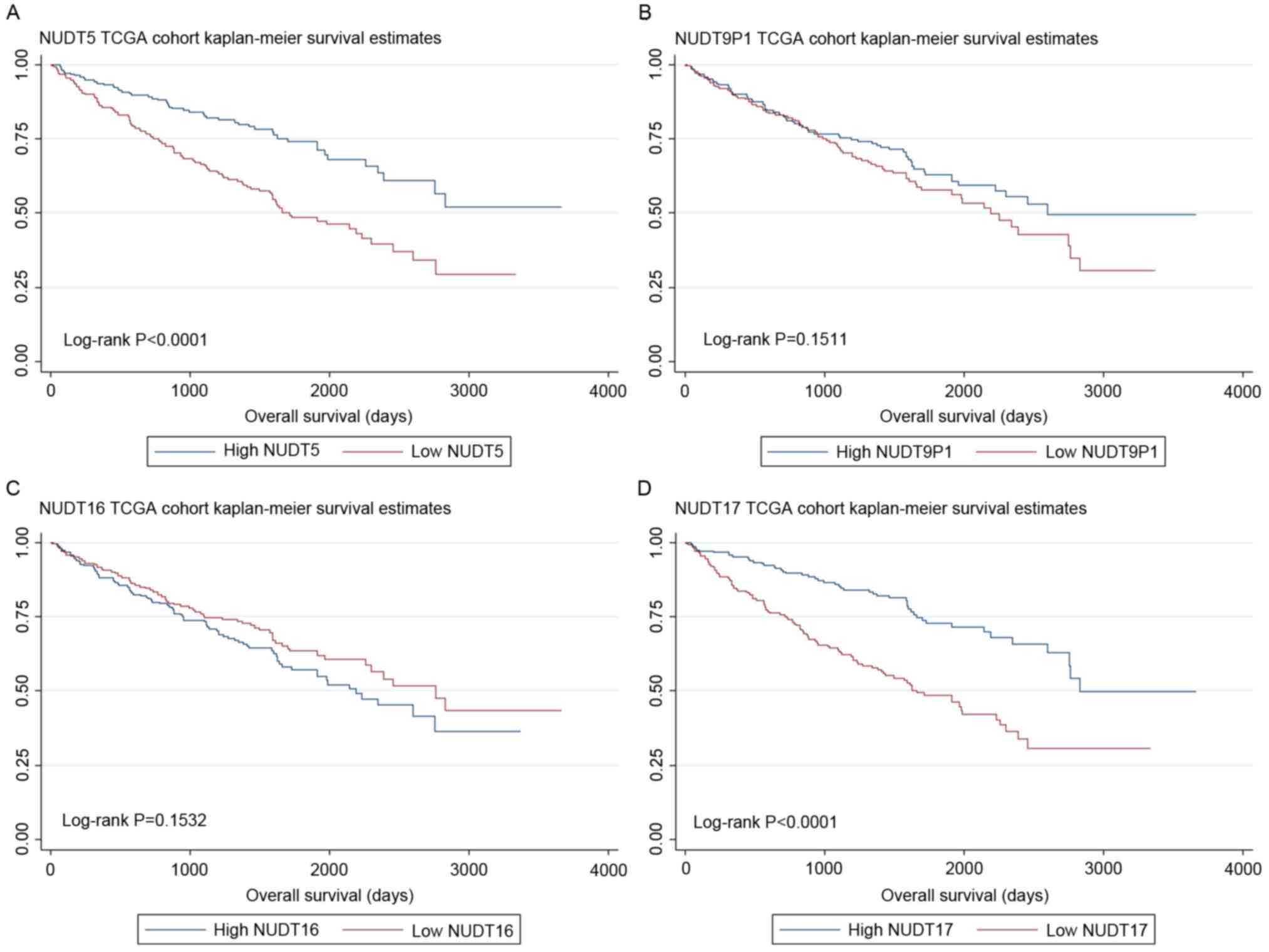
High expression of NUDT5 and NUDT17
were associated with better prognosis and longer OS in the TCGA
cohort
Further study of NUDT5, NUDT9P1, NUDT16 and NUDT17
expression revealed that they were normally distributed (data not
shown), so TCGA cohort was divided into low and high expression
groups according to the median expression level. As a result,
higher NUDT5 (P<0.0001) and NUDT17 (P<0.0001) expression was
associated with better prognosis for OS, whereas high levels of
NUDT9P1 (P=0.151) and NUDT16 (P=0.153) expression was not
associated with OS prognosis (Fig.
1).
In multivariate logistic regression analysis of
factors that could affect the expression of NUDT5, NUDT9P1, NUDT16
and NUDT17, tumor grade was significantly associated with NUDT5
(P=0.006) and NUDT17 (P=0.002) expression, while tumor stage was
also significantly associated with NUDT5 (P=0.001) and NUDT17
(P=0.007) expression (Table
III).
 | Table III.Multivariate logistic regression
analysis of factors that might affect the expression of NUDT5,
NUDT9P1, NUDT16 and NUDT17 in The Cancer Genome Atlas cohort with
clear cell renal cell carcinoma. |
Table III.
Multivariate logistic regression
analysis of factors that might affect the expression of NUDT5,
NUDT9P1, NUDT16 and NUDT17 in The Cancer Genome Atlas cohort with
clear cell renal cell carcinoma.
|
| NUDT5 | NUDT9P1 | NUDT16 | NUDT17 |
|---|
|
|
|
|
|
|
|---|
| Variables | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | P-value |
|---|
| Age | 1.009 | 0.992–1.026 | 0.288 | 0.984 | 0.968–1.000 | 0.055 | 1.008 | 0.991–1.023 | 0.374 | 0.994 | 0.978–1.011 | 0.507 |
| Stage | 1.409 | 1.148–1.732 | 0.001 | 1.155 | 0.947–1.409 | 0.155 | 0.926 | 0.760–1.128 | 0.446 | 1.318 | 1.078–1.613 | 0.007 |
| Grade | 1.568 | 1.138–2.160 | 0.006 | 0.800 | 0.590–1.085 | 0.152 | 1.029 | 0.761–1.390 | 0.855 | 1.637 | 1.197–2.239 | 0.002 |
| Hb | 0.898 | 0.588–1.370 | 0.617 | 0.734 | 0.489–1.103 | 0.137 | 1.171 | 0.783–1.752 | 0.442 | 0.961 | 0.633–1.459 | 0.852 |
| WBC | 0.937 | 0.604–1.453 | 0.771 | 0.804 | 0.529–1.222 | 0.307 | 1.302 | 0.859–1.973 | 0.213 | 0.977 | 0.635–1.502 | 0.916 |
| PLT | 1.563 | 0.013–0.359 | 0.085 | 1.221 | 0.765–1.949 | 0.403 | 0.942 | 0.593–1.496 | 0.799 | 0.742 | 0.458–1.201 | 0.226 |
To understand the different expression of NUDT
family between patients with ccRCC and normal population further,
the present study analyzed the expression of NUDT family in 70
couples of paired patients. If deviations in NUDT expression
between couples fitted a normal distribution, paired student
t-tests were performed; if not, Wilcoxon signed-rank test was
performed. Using a paired Student's t-test, the expression of
NUDT3, NUDT4, NUDT6, NUDY7, NUDT9SP1, NUDT12, NUDT13, NUDT15,
NUDT16 and NUDT16SP1 was found to be significantly different
between patients with ccRCC and paired healthy individuals, whereas
differences in the expression of NUDT17 was not statistically
significant. Using a Wilcoxon signed-rank test, expression of
NUDT1, NUDT8, NUDT9, NUDT10, NUDT11, NUDT16L1, NUDT18 and NUDT21
were significantly different between patients with ccRCC and paired
healthy individuals, whereas expression of NUDT2, NUDT5, NUDt14,
NUDT19 and NUDT22 did not differ significantly (Table IV).
 | Table IV.Expression of NUDT family genes in 70
patients and paired healthy individuals in The Cancer Genome Atlas
cohort. |
Table IV.
Expression of NUDT family genes in 70
patients and paired healthy individuals in The Cancer Genome Atlas
cohort.
| Variables | P-value | Statistical
test | 95% CI |
|---|
| NUDT9P1 | <0.001 | Paired Student
t-test | 0.364–0.677 |
| NUDT16L1 | <0.001 | Wilcoxon rank-sum
test |
|
| NUDT12 | <0.001 | Paired Student
t-test | 0.467–0.818 |
| NUDT10 | <0.001 | Wilcoxon rank-sum
test |
|
| NUDT17 | 0.117 | Paired Student
t-test | −0.490–0.056 |
| NUDT14 | 0.301 | Wilcoxon rank-sum
test |
|
| NUDT15 | <0.001 | Paired Student
t-test | 0.110–0.338 |
| NUDT18 | <0.001 | Wilcoxon rank-sum
test |
|
| NUDT19 | 0.859 | Wilcoxon rank-sum
test |
|
| NUDT11 | <0.001 | Wilcoxon rank-sum
test |
|
| NUDT13 | <0.001 | Paired Student
t-test | 0.199–0.531 |
| NUDT1 | <0.001 | Wilcoxon rank-sum
test |
|
| NUDT2 | 0.850 | Wilcoxon rank-sum
test |
|
| NUDT3 | 0.001 | Paired Student
t-test | 0.0742–0.279 |
| NUDT4 | <0.001 | Paired Student
t-test | 1.638–2.140 |
| NUDT5 | 0.149 | Wilcoxon rank-sum
test |
|
| NUDT6 | <0.001 | Paired Student
t-test | 0.951–1.344 |
| NUDT7 | <0.001 | Paired Student
t-test | 0.667–0.982 |
| NUDT9 | <0.001 | Wilcoxon rank-sum
test |
|
| NUDT8 | 0.002 | Wilcoxon rank-sum
test |
|
| NUDT22 | 0.160 | Wilcoxon rank-sum
test |
|
| NUDT21 | <0.001 | Wilcoxon rank-sum
test |
|
| NUDT16P1 | <0.001 | Paired Student
t-test | 0.322–0.633 |
| NUDT16 | <0.001 | Paired Student
t-test | 0.559–0.821 |
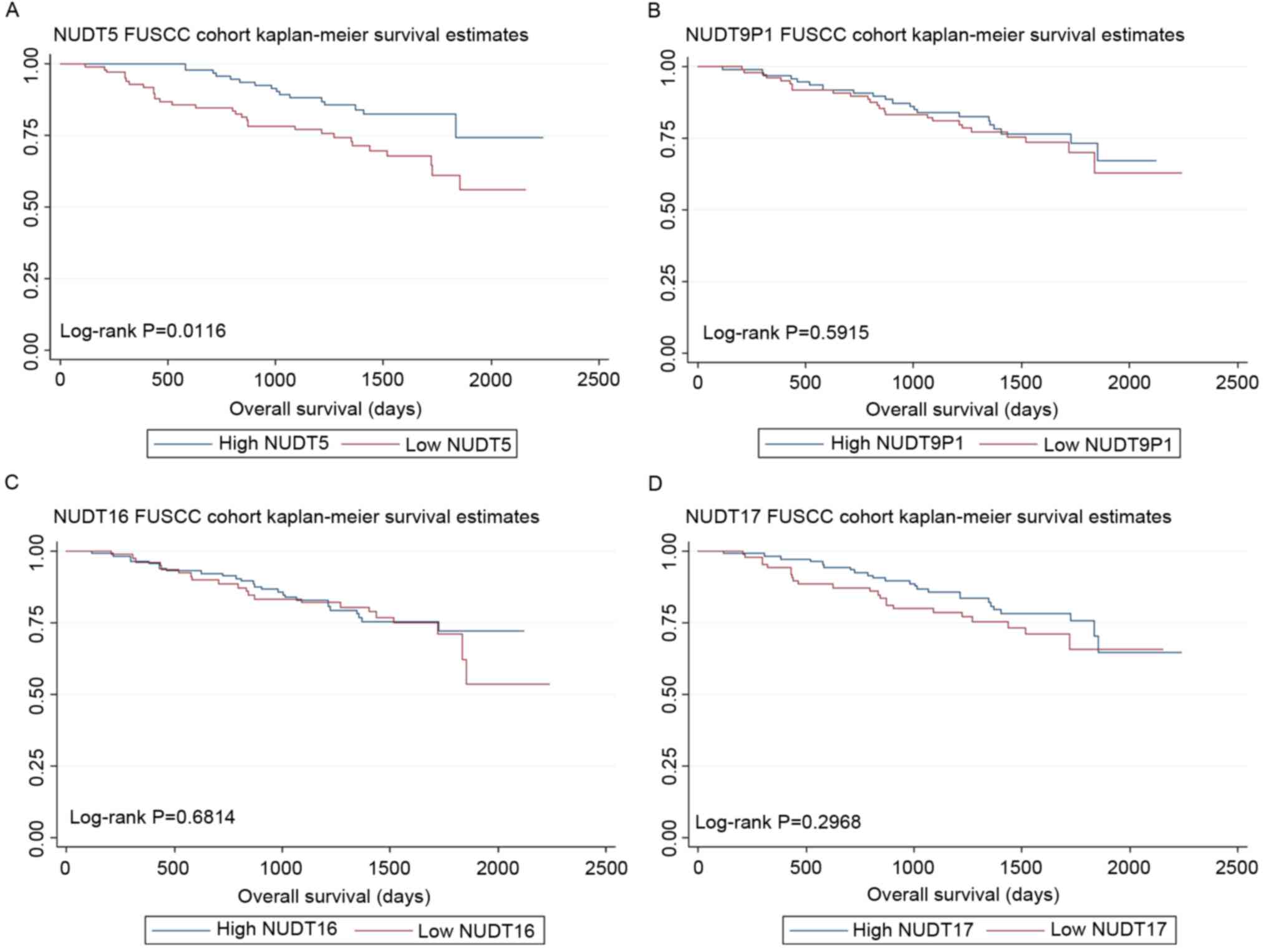
NUDT5 expressions were prognostic
factors for OS in the FUSCC cohort
NUDT5, NUDT9P1, NUDT16 and NUDT17 expression was
validated in the FUSCC cohort. This cohort was then divided into
low- and high-expression groups according to the median expression
level. As the expression level of genes was based on the relative
values of PCR results, patients were grouped by Δ-Ct (cycle
threshold). Δ-Ct=Ct (target genes)-Ct (reference genes). The median
Δ-Ct value of NUDT5, NUDT9P1, NUDT16 and NUDT17 were 8.29, 3.90,
7.32 and 4.67, respectively. As a result, low NUDT5 expression was
associated with poor OS (log-rank test, P=0.0116), although the
level of NUDT9P1 (log-rank test, P=0.5915), NUDT16 (log-rank test,
P=0.6814) and NUDT17 (log-rank test, P=0.2968) expression was not
associated with OS. The Kaplan-Meier curves are shown in Fig. 2.
To understand the factors that may affect the
expression of NUDT5, NUDT9P1, NUDT16 and NUDT17 in the FUSCC cohort
further, multivariate logistic regression analysis with the same
parameters including age, stage, grade, hemoglobin level; white
blood cells level and platelets level was performed. In the FUSCC
cohort, tumor grade was significantly associated with the NUDT5
expression level (P=0.016) expression, whereas other parameters
were not significantly associated with the expression of NUDTs
(Table V).
 | Table V.Multivariate logistic regression
analysis of factors that might affect the expression of NUDT5,
NUDT9P1, NUDT16 and NUDT17 in the Fudan University Shanghai Cancer
Center cohort with clear cell renal cell carcinoma. |
Table V.
Multivariate logistic regression
analysis of factors that might affect the expression of NUDT5,
NUDT9P1, NUDT16 and NUDT17 in the Fudan University Shanghai Cancer
Center cohort with clear cell renal cell carcinoma.
|
| NUDT5 | NUDT9P1 | NUDT16 | NUDT17 |
|---|
|
|
|
|
|
|
|---|
| Variables | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | P-value |
|---|
| Age | 0.992 | 0.959–1.027 | 0.667 | 0.984 | 0.950–1.018 | 0.355 | 0.978 | 0.944–1.013 | 0.225 | 0.992 | 0.958–1.027 | 0.659 |
| Stage | 0.663 | 0.409–1.077 | 0.097 | 1.380 | 0.868–2.197 | 0.174 | 1.000 | 0.616–1.623 | 0.998 | 1.539 | 0.961–2.462 | 0.073 |
| Grade | 2.132 | 1.150–3.951 | 0.016 | 0.770 | 0.437–1.356 | 0.366 | 0.789 | 0.438–1.421 | 0.430 | 1.400 | 0.794–2.470 | 0.245 |
| Hb | 0.984 | 0.958–1.011 | 0.243 | 1.023 | 0.995–1.051 | 0.104 | 1.011 | 0.984–1.038 | 0.420 | 1.018 | 0.991–1.046 | 0.198 |
| WBC | 0.995 | 0.987–1.003 | 0.222 | 0.999 | 0.993–1.005 | 0.723 | 0.993 | 0.981–1.005 | 0.275 | 0.997 | 0.992–1.003 | 0.412 |
| PLT | 1.002 | 0.996–1.007 | 0.561 | 0.998 | 0.992–1.003 | 0.477 | 1.001 | 0.995–1.007 | 0.672 | 1.004 | 0.998–1.009 | 0.192 |
Discussion
To the best of our knowledge, the present study
demonstrates that expression of the NUDT family of genes is
associated with the OS of patients with ccRCC. Members of this
family, particularly NUDT5, NUDT9P1, NUDT16 and NUDT17, may be
independent prognostic factors for OS in patients with ccRCC.
The present study demonstrates that the NUDT family
may have important roles in suppressing the progression of ccRCC.
NUDT5, NUDT9P1, NUDT16 and NUDT17 expression were independent
prognostic factors for OS in patients with ccRCC. Reduced
expression of NUDT5 and NUDT17 was associated with poor prognosis
and decreased OS time. Expression of NUDT5 is closely associated
with the prognosis of patients with ccRCC. Additionally, Fuhrman
grade and TNM stage were significantly associated with NUDT5 and
NUDT17 expression. However, upon statistical analysis of 70 paired
patients with ccRCC and healthy individuals, there was no
significant difference in expression of NUDT5 and NUDT17. This may
be because: i) The number of patients included in the paired study
was not large enough; and ii) in analysis of NUDT5, the Wilcoxon
signed-rank test was used, which has a low power and thus may
affect the outcome of the analysis. However, in patients in TCGA
database, NUDT5 and NUDT17 remained good indicators of prognosis.
To further verify the accuracy of NUDT as a ccRCC prognostic
marker, FUSCC patient specimens were tested. Low NUDT5 expression
was associated with OS rates in the FUSCC cohort. Even though no
statistical difference existed between low and high NUDT17
expression groups in the FUSCC cohort, NUDT5 remains a good
prognostic indicator, due to demographic difference between these
two studies. All patients in the present study were Asian, while
the majority of the patients in the TCGA cohort were Caucasian or
of African descent.
The human genome has 24 NUDT hydrolase genes and at
least 5 pseudogenes (18). NUDT genes
are associated with metabolic reprogramming and mutagenesis.
Previous studies have partially revealed their functions, even
though the role they serve in tumorigenesis is poorly understood
(16,17).
NUDT5 is an antimutator candidate; this protein was
originally characterized as an ADP sugar hydrolase, which
corresponds to the high-Km ADP Ribose-II isolated from
tissues. ADP Ribose is a member of a family of proteins involved in
a number of cellular processes such as DNA repair, genomic
stability and programmed cell death (19,20). In
experiments in vitro, NUDT5 suppressed the increased
mutation rate of cancer cells and may act in concert with NUDT1 or
NUDT15 in antimutagenesis (19,20). NUDT5
may also prevent transcriptional errors and mistranslation. Prior
studies (19–21) also found that lowered NUDT5 expression
led to cell cycle inhibition in HeLa cells (21). Further studies indicated that the
NUDT5 protein may have notable roles in regulating the G1-S
transition in mammalian cells (22–24).
Nudix hydrolase 9 pseudogene 1 (NUDT9P1) is located
in the 5-HT receptor 7, adenylate cyclase-coupled (HTR7) gene,
which is associated with the response to iloperidone. However, the
role of NUDT9P1 in healthy or tumor cells remains unknown (25).
NUDT16 is a ‘housecleaning’ enzyme that removes
inosine diphosphate from the nucleotide pool. Studies have revealed
that NUDT16 forms a dimer, which generates a positively charged
trench to accommodate substrate binding (26). NUDT16 may be involved in regulating
ribosome biogenesis by altering the stability of U8 small nucleolar
RNA and other guide RNAs (26,27).
Studies have revealed that NUDT16 may interact with a nuclear
protein phosphatase, possibly in a complex with small nuclear
riboprotein components (28,29).
At the time of writing, NUDT17 remains an
uncharacterized protein, with no known function. NUDT17 may be
bi-functional and possess mRNA de-capping activity in cells, in
addition to its reported activities on nucleotide containing
molecules (16).
Little is known about the NUDT family of genes in
the field of oncology. NUDT1 and NUDT15 are expressed in
RAS-dependent types of cancer (30,31). Loss
of NUDT1 function impaired growth of KRAS proto-oncogene,
GTPase-positive tumor cells. NUDT1 overexpression mitigated
sensitivity towards certain experimental small molecules, including
the NUDT1 inhibitor SCH51344 (30–32).
However, the association between NUDT family and tumorigenesis were
not clear and studies about their role in renal cancer are rare
(30–32).
The present study confirmed the role of the NUDT
family of genes in patients with ccRCC, identifying NUDT5 may
inform on patient prognosis. Limitations of the present study are:
i) All of the patients that were included in the present study were
from Fudan University Shanghai Cancer Center with excellent
follow-up, and patients from other centers were not included; ii)
all patient tissue specimens in the present study came from
patients who suitable to surgery so it is possible that the results
will not apply to people who were not suitable for surgery; and
iii) the number of patients who participated in the study was
low.
The present study indicated the presence of an
association between ccRCC outcome and NUDT gene family expression;
however, the underlying mechanism has yet to be elucidated. The
present study may have revealed novel ccRCC biomarkers or
therapeutic targets; as such, further study is urged.
NUDT5 expression was identified as an independent
prognostic factor for OS time of ccRCC in the present study: Low
NUDT5 expression was associated with low OS time and tumor grade
was significantly associated with NUDT5 expression. NUDT5 could
therefore act as a tool to reveal further prognostic genes in
ccRCC.
Acknowledgements
The present study was supported in part by the
grants for International Cooperation and Exchange of Science and
Technology Commission of Shanghai Municipality (no. 12410709300),
from the Guide Project of Science and Technology Commission of
Shanghai Municipality (no. 124119a7300), and from the Outstanding
Young Talent Training Plan of Shanghai Municipal Commission of
Health and Family Planning (no. XYQ2013102).
References
|
1
|
Ljungberg B, Campbell SC, Choi HY, Jacqmin
D, Lee JE, Weikert S and Kiemeney LA: The epidemiology of renal
cell carcinoma. Eur Urol. 60:615–621. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ridge CA, Pua BB and Madoff DC:
Epidemiology and staging of renal cell carcinoma. Semin Intervent
Radiol. 31:3–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shuch B, Ricketts CJ, Vocke CD, Komiya T,
Middelton LA, Kauffman EC, Merino MJ, Metwalli AR, Dennis P and
Linehan WM: Germline PTEN mutation Cowden syndrome: An
underappreciated form of hereditary kidney cancer. J Urol.
190:1990–1998. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chow WH, Dong LM and Devesa SS:
Epidemiology and risk factors for kidney cancer. Nat Rev Urol.
7:245–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ljungberg B, Cowan NC, Hanbury DC, Hora M,
Kuczyk MA, Merseburger AS, Patard JJ, Mulders PF and Sinescu IC:
European Association of Urology Guideline Group: EAU guidelines on
renal cell carcinoma: The 2010 update. Eur Urol. 58:398–406. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Maher ER: Genomics and epigenomics of
renal cell carcinoma. Semin Cancer Biol. 23:10–17. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Linehan WM, Srinivasan R and Schmidt LS:
The genetic basis of kidney cancer: A metabolic disease. Nat Rev
Urol. 7:277–285. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bessman MJ, Frick DN and O'Handley SF: The
MutT proteins or ‘Nudix’ hydrolases, a family of versatile, widely
distributed, ‘housecleaning’ enzymes. J Biol Chem. 271:25059–25062.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ito R, Hayakawa H, Sekiguchi M and
Ishibashi T: Multiple enzyme activities of Escherichia coli MutT
protein for sanitization of DNA and RNA precursor pools.
Biochemistry. 44:6670–6674. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu W, Jones CR, Dunn CA and Bessman MJ:
Gene ytkD of Bacillus subtilis encodes an atypical nucleoside
triphosphatase member of the Nudix hydrolase superfamily. J
Bacteriol. 186:8380–8384. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fisher DI, Cartwright JL, Harashima H,
Kamiya H and McLennan AG: Characterization of a nudix hydrolase
from Deinococcus radiodurans with a marked specificity for
(deoxy)ribonucleoside 5′-diphosphates. BMC Biochem. 5:72004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Safrany ST, Caffrey JJ, Yang X, Bembenek
ME, Moyer MB, Burkhart WA and Shears SB: A novel context for the
‘MutT’ module, a guardian of cell integrity, in a diphosphoinositol
polyphosphate phosphohydrolase. EMBO J. 17:6599–6607. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fujikawa K, Kamiya H, Yakushiji H, Fujii
Y, Nakabeppu Y and Kasai H: The oxidized forms of dATP are
substrates for the human MutT homologue, the hMTH1 protein. J Biol
Chem. 274:18201–18205. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kamiya H, Yakushiji H, Dugué L, Tanimoto
M, Pochet S, Nakabeppu Y and Harashima H: Probing the substrate
recognition mechanism of the human MTH1 protein by nucleotide
analogs. J Mol Biol. 336:843–850. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Caffrey JJ, Safrany ST, Yang X and Shears
SB: Discovery of molecular and catalytic diversity among human
diphosphoinositol-polyphosphate phosphohydrolases. An expanding
Nudt family. J Biol Chem. 275:12730–12736. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McLennan AG, Cartwright JL and Gasmi L:
The human NUDT family of nucleotide hydrolases. Enzymes of diverse
substrate specificity. Adv Exp Med Biol. 486:115–118. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kamiya H, Cadena-Amaro C, Dugué L,
Yakushiji H, Minakawa N, Matsuda A, Pochet S, Nakabeppu Y and
Harashima H: Recognition of nucleotide analogs containing the
7,8-dihydro-8-oxo structure by the human MTH1 protein. J Biochem.
140:843–849. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gasmi L, Cartwright JL and McLennan AG:
Cloning, expression and characterization of YSA1H, a human
adenosine 5′-diphosphosugar pyrophosphatase possessing a MutT
motif. Biochem J. 344:331–337. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang H, Slupska MM, Wei YF, Tai JH, Luther
WM, Xia YR, Shih DM, Chiang JH, Baikalov C, Fitz-Gibbon S, et al:
Cloning and characterization of a new member of the Nudix
hydrolases from human and mouse. J Biol Chem. 275:8844–8853. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang LQ, Dai DP, Gan W, Takagi Y,
Hayakawa H, Sekiguchi M and Cai JP: Lowered nudix type 5 (NUDT5)
expression leads to cell cycle retardation in HeLa cells. Mol Cell
Biochem. 363:377–384. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fujiwara S, Sawada K and Amisaki T:
Molecular dynamics study on conformational differences between dGMP
and 8-oxo-dGMP: Effects of metal ions. J Mol Graph Model.
51:158–167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Arimori T, Tamaoki H, Nakamura T, Kamiya
H, Ikemizu S, Takagi Y, Ishibashi T, Harashima H, Sekiguchi M and
Yamagata Y: Diverse substrate recognition and hydrolysis mechanisms
of human NUDT5. Nucleic Acids Res. 39:8972–8983. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zha M, Guo Q, Zhang Y, Yu B, Ou Y, Zhong C
and Ding J: Molecular mechanism of ADP-ribose hydrolysis by human
NUDT5 from structural and kinetic studies. J Mol Biol. 379:568–578.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lavedan C, Licamele L, Volpi S, Hamilton
J, Heaton C, Mack K, Lannan R, Thompson A, Wolfgang CD and
Polymeropoulos MH: Association of the NPAS3 gene and five other
loci with response to the antipsychotic iloperidone identified in a
whole genome association study. Mol Psychiatry. 14:804–819. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu G, Zhang J, Li Y, Li Z, Zhang N, Xu X,
Wang T, Guan Z, Gao GF and Yan J: hNUDT16: A universal decapping
enzyme for small nucleolar RNA and cytoplasmic mRNA. Protein Cell.
2:64–73. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Y, Song M and Kiledjian M: Differential
utilization of decapping enzymes in mammalian mRNA decay pathways.
RNA. 17:419–428. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Trésaugues L, Lundbäck T, Welin M, Flodin
S, Nyman T, Silvander C, Gräslund S and Nordlund P: Structural
basis for the specificity of human NUDT16 and its regulation by
inosine monophosphate. PLoS One. 10:e01315072015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Abolhassani N, Iyama T, Tsuchimoto D,
Sakumi K, Ohno M, Behmanesh M and Nakabeppu Y: NUDT16 and ITPA play
a dual protective role in maintaining chromosome stability and cell
growth by eliminating dIDP/IDP and dITP/ITP from nucleotide pools
in mammals. Nucleic Acids Res. 38:2891–2903. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dallosso AR, Dolwani S, Jones N, Jones S,
Colley J, Maynard J, Idziaszczyk S, Humphreys V, Arnold J,
Donaldson A, et al: Inherited predisposition to colorectal adenomas
caused by multiple rare alleles of MUTYH but not OGG1, NUDT1, NTH1
or NEIL 1, 2 or 3. Gut. 57:1252–1255. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cho WC, Chow AS and Au JS: MiR-145
inhibits cell proliferation of human lung adenocarcinoma by
targeting EGFR and NUDT1. RNA Biol. 8:125–131. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Garre P, Briceño V, Xicola RM, Doyle BJ,
de la Hoya M, Sanz J, Llovet P, Pescador P, Puente J, Díaz-Rubio E,
et al: Analysis of the oxidative damage repair genes NUDT1, OGG1,
and MUTYH in patients from mismatch repair proficient HNPCC
families (MSS-HNPCC). Clin Cancer Res. 17:1701–1712. 2011.
View Article : Google Scholar : PubMed/NCBI
|