Introduction
Colorectal cancer (CRC) is the third most common
malignancy worldwide and the third leading cause of
cancer-associated mortality in the United States (1,2). It is
estimated that ~1.3 million new cases are diagnosed and ~0.7
million people succumb to the disease each year worldwide (3). According to recent statistics, it was
estimated that 134,490 new cases and 49,190 fatalities would occur
in America in 2016 (1). The incidence
and mortality rates for men and women have improved in the last
decades as a result of advances in screening and clinical treatment
(4,5).
Metastasis, the most common cause of cancer-associated mortality,
is a multi-step process through which tumor cells spread from their
primary site and form secondary growths at a distance (6). Metastasis is among the six initially
described hallmarks of cancer and is a major cause of
CRC-associated mortality (7,8). The 5-year survival rate of early-stage
CRC ranges between 60 and 95%; however, for patients with
metastatic tumors, the survival rate ranges from 10 to 35%
(9–11). Despite improvements in diagnosis and
treatment, ~90% of CRC-associated mortalities are due to metastases
and it has been estimated that ~50% of all patients diagnosed with
CRC eventually succumb to metastatic disease (12,13).
Therefore, it is critical to investigate the mechanism of
metastasis in CRC and identify novel molecular therapeutic and
prognostic targets. To date, some progress has been made.
Epithelial-mesenchymal transition was demonstrated to serve a
pivotal and intricate role in promoting CRC metastasis (6,14). In
addition, certain genes and proteins were found to be associated
with CRC metastasis, including semaphoring 3F, stromal interaction
molecule 1, forkhead box C2 and hes family BHLH transcription
factor 1 (14–17), as well as Cyclin b1, Angiopoietin-like
4 and p21-activated kinase 1 and 4 (18–20).
However, one study demonstrated that metastasis occurs through a
multistep cascade of events, but that it was inefficient as a whole
process (8). Even though mutations
associated with metastasis have been investigated in the past, only
a limited number of such genetic alterations have been identified
and the underlying molecular mechanism remains unclear (21). The present study analyzed the microRNA
(miR/miRNA) and mRNA expression profiles of CRC samples in order to
investigate the mechanism of metastasis in CRC and identify novel
molecular biomarkers.
Materials and methods
mRNA and miRNA expression data
The mRNA and miRNA expression profiles of the
GSE2509 (22) and GSE56350 (23) datasets were obtained from the Gene
Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/). GSE2509 contains the mRNA
profile of 3 primary CRC samples and 3 samples with lymph node
metastasis. These data were analyzed with the GPL96 [HG-U133A]
Affymetrix Human Genome U133A Array platform version 2.0
(Affymetrix; Fisher Scientific, Inc., Waltham, MA, USA).
Additionally, 46 primary CRC samples and 43 CRC samples with lymph
node metastasis were obtained from the miRNA profile of GSE56350.
Detection of the miRNA profile was performed using the PL16744
OSU-CCC Human and Mouse MicroRNA Microarray platform version 4.0
(Comprehensive Cancer Center, The Ohio State University, Columbus,
OH, USA).
Data processing and differential
expression analysis
The mRNA profile data were converted into
recognizable format in R and then were normalized using the Robust
Multi-Array Average algorithm from Affy version 1.40.0
package (http://www.bioconductor.org/packages/2.13/bioc/html/affy.html)
(24). For the miRNA profile, the
original expression value matrix was obtained and normalization was
conducted using the preprocessCore function package version 3.5
(http://www.bioconductor.org/packages/release/bioc/html/preprocessCore.html)
(25). Subsequently, the
differentially expressed genes (DEGs) and differentially expressed
miRNAs (DEMs) were identified in the CRC samples with lymph node
metastasis compared with the primary CRC samples using the limma
version 3.18.13 software package (http://www.bioconductor.org/packages/2.13/bioc/html/limma.html)
(26). P<0.05 and |log2
(fold-change)| >0.2 were used as threshold criteria.
Functional and pathway enrichment
analysis of DEGs
The Database for Annotation, Visualization and
Integrated Discovery version 6.8 (https://david.ncifcrf.gov/) (27) was used to perform Gene Ontology (GO)
and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment analysis of DEGs. GO terms and KEGG pathways with
P<0.05 were selected.
Construction of the miRNA-gene
network
The miRWalk version 2.0 database (mirwalk.uni-hd.de/) is a publicly available
comprehensive resource containing the predicted and the
experimentally validated miRNA-target interaction pairs (28). The DEM-associated predicted target
genes were selected when they were included in at least four out of
five databases (miRanda-rel2010, miRDB version 4.0, miRWalk version
2.0, RNA22 version 2.0 and TargetScan version 6.2). Subsequently,
the overlapping target genes were identified and the miRNA-gene
pairs and DEMs were selected. The miRNA-gene network was formed and
visualized using the Cytoscape version 3.5.1 (29) software (http://www.cytoscape.org/download.php).
Functional enrichment analysis of
predicted target genes regulated by DEMs
The clusterProfiler version 3.5 (http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html),
an R package, was used to perform biological-term classification
and enrichment analysis of gene clusters (30). Following the identification of the
predicted target genes regulated by DEMs, enrichment analysis was
performed and the enriched pathways with a P-value of <0.05 were
selected.
Results
Differentially expressed mRNAs and
differentially expressed miRNAs
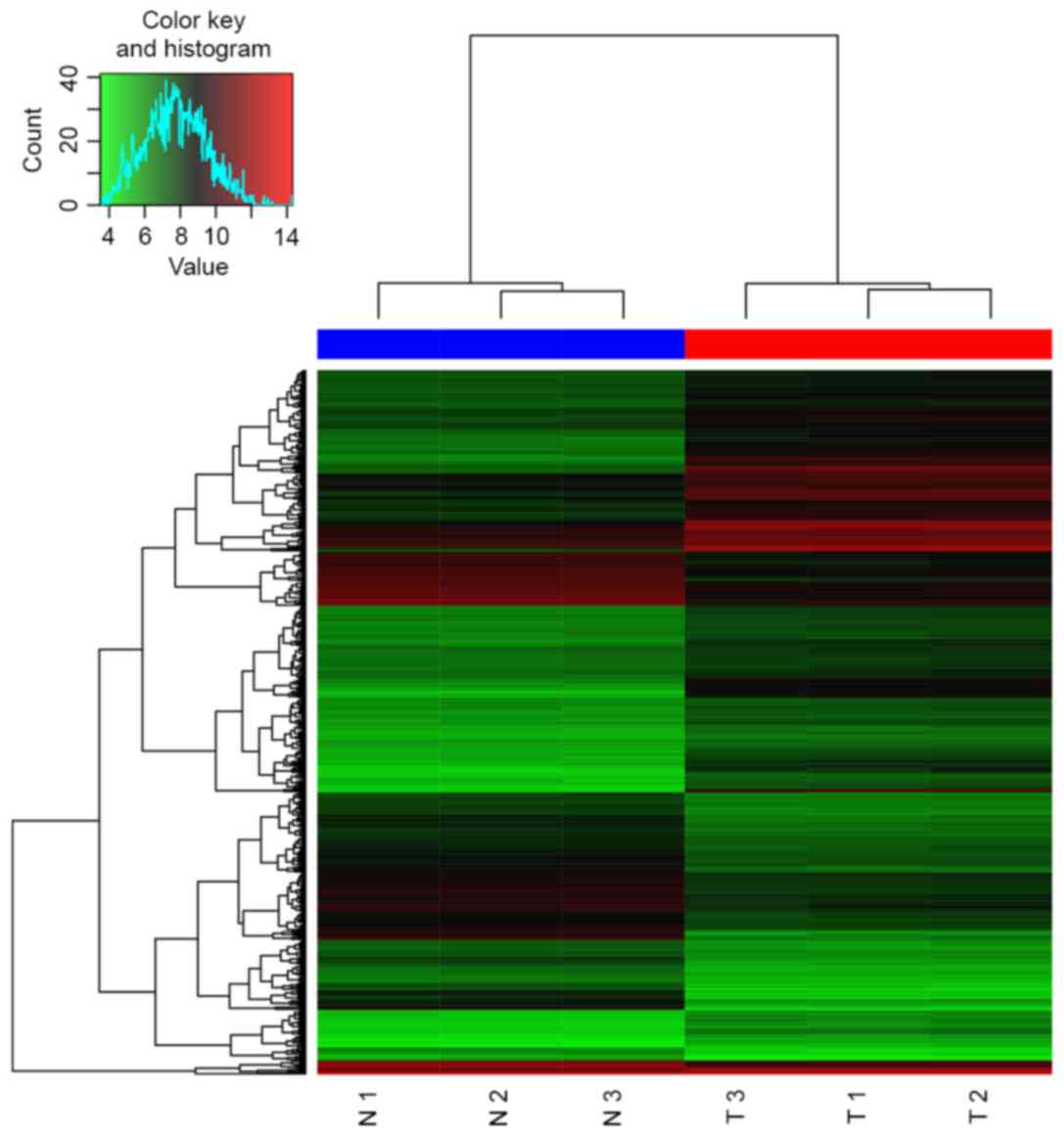
A total of 544 DEGs (227 upregulated and 317
downregulated) were identified, and the heat map of hierarchical
clustering is presented in Fig. 1.
Additionally, 42 DEMs (25 upregulated and 17 downregulated) were
identified. The 20 most significant DEGs and the 20 most
significant DEMs are presented in Tables
I and II, respectively.
 | Table I.20 most significant differentially
expressed genes in colorectal cancer samples with lymph node
metastasis. |
Table I.
20 most significant differentially
expressed genes in colorectal cancer samples with lymph node
metastasis.
| Gene | P-value | log2
FC |
|---|
| RNF128 |
4.07×10−15 | 4.435925 |
| CNN3 |
6.05×10−15 | 4.696778 |
| WNT5A |
8.42×10−15 | −5.04177 |
| TOX3 |
9.65×10−15 | 5.640899 |
| FZD10 |
1.13×10−14 | 3.438707 |
| IGFBP3 |
3.26×10−14 | −4.23336 |
| AKR1C3 |
4.50×10−14 | 4.643379 |
| TRIP6 |
5.04×10−14 | −3.79486 |
| NPC2 |
5.37×10−14 | −3.86253 |
| KRT13 |
8.49×10−14 | −5.41489 |
| KRT23 |
8.91×10−14 | −4.84643 |
| NGFR |
9.23×10−14 | −3.8684 |
| KRT81 |
9.38×10−14 | −3.54854 |
| CXCR4 |
1.25×10−13 | −3.41813 |
| ANXA6 |
1.29×10−13 | −3.29244 |
| MSX1 |
1.64×10−13 | −3.64506 |
| EFNB2 |
1.76×10−13 | 3.675191 |
| SLC2A3 |
1.77×10−13 | −4.2354 |
| IGFBP7 |
2.18×10−13 | −3.27699 |
| GNG11 |
2.77×10−13 | −5.77814 |
 | Table II.20 most significant differentially
expressed miRNAs in colorectal cancer samples with lymph node
metastasis. |
Table II.
20 most significant differentially
expressed miRNAs in colorectal cancer samples with lymph node
metastasis.
| miRNA | P-value | log2
FC |
|---|
|
hsa-miR-342-3p |
5.2×10−10 | 1.3803 |
|
hsa-miR-150 |
1.31×10−9 | 1.5415 |
|
hsa-miR-155 |
2.73×10−7 | 1.5434 |
|
hsa-miR-92b |
6.19×10−7 | −1.6535 |
|
hsa-miR-375 |
4.04×10−6 | −1.8421 |
|
hsa-miR-142-5p |
3.72×10−6 | 1.752 |
|
hsa-miR-453 |
8.88×10−6 | −1.1169 |
|
hsa-miR-622 |
1.47×10−5 | −1.4782 |
|
hsa-miR-595 |
6.48×10−5 | −1.0289 |
|
hsa-miR-629 |
8.46×10−5 | −1.1146 |
|
hsa-mir-621 |
1.04×10−4 | −1.2234 |
|
hsa-miR-26a |
1.19×10−4 | 2.4369 |
|
hsa-miR-146a |
1.51×10−4 | 2.1241 |
|
hsa-miR-26b |
1.56×10−4 | 2.7188 |
|
hsa-miR-200b |
2.05×10−4 | −1.1071 |
|
hsa-miR-146b-5p |
2.18×10−4 | 1.9222 |
|
hsa-miR-107 |
2.91×10−4 | 2.1441 |
|
hsa-miR-560 |
3.29×10−4 | −1.4887 |
|
hsa-mir-766 |
3.33×10−4 | −1.1362 |
|
hsa-miR-103 |
4.97×10−4 | 2.1098 |
Enriched GO terms and KEGG pathways of
differentially expressed mRNAs
DEGs were enriched in 320 GO terms and 11 KEGG
pathways. The 10 most significant enriched GO terms and the
enriched KEGG pathways are presented in Tables III and IV, respectively.
 | Table III.10 most significant enriched GO terms
of differentially expressed microRNAs. |
Table III.
10 most significant enriched GO terms
of differentially expressed microRNAs.
| Category | GO ID | GO name | Count | P-value |
|---|
| BP | GO:0001944 | Vasculature
development | 29 |
1.30×10−8 |
| BP | GO:0001568 | Blood vessel
development | 28 |
3.08×10−8 |
| BP | GO:0016477 | Cell migration | 27 |
1.22×10−6 |
| BP | GO:0048514 | Blood vessel
morphogenesis | 22 |
5.54×10−6 |
| BP | GO:0051674 | Localization of
cell | 27 |
8.67×10−6 |
| BP | GO:0048870 | Cell motility | 27 |
8.67×10−6 |
| BP | GO:0042127 | Regulation of cell
proliferation | 50 |
9.42×10−6 |
| BP | GO:0051094 | Positive regulation
of developmental process | 25 |
1.40×10−5 |
| BP | GO:0006928 | Cell motion | 35 |
1.46×10−5 |
| BP | GO:0045597 | Positive regulation
of cell differentiation | 22 |
1.95×10−5 |
 | Table IV.Enriched KEGG pathways of
differentially expressed microRNAs. |
Table IV.
Enriched KEGG pathways of
differentially expressed microRNAs.
| Category | Pathway name | Count | P-value |
|---|
| KEGG_PATHWAY | hsa05200: Pathways
in cancer | 24 | 0.00217 |
| KEGG_PATHWAY | hsa04360: Axon
guidance | 12 | 0.008095 |
| KEGG_PATHWAY | hsa04310: Wnt
signaling pathway | 13 | 0.009973 |
| KEGG_PATHWAY | hsa05222: Small
cell lung cancer | 9 | 0.012226 |
| KEGG_PATHWAY | hsa04115: p53
signaling pathway | 8 | 0.012507 |
| KEGG_PATHWAY | hsa05217: Basal
cell carcinoma | 7 | 0.015538 |
| KEGG_PATHWAY | hsa04916:
Melanogenesis | 9 | 0.030049 |
| KEGG_PATHWAY | hsa04060:
Cytokine-cytokine receptor interaction | 17 | 0.033293 |
| KEGG_PATHWAY | hsa05210:
Colorectal cancer | 8 | 0.035683 |
| KEGG_PATHWAY | hsa04512:
ECM-receptor interaction | 8 | 0.035683 |
| KEGG_PATHWAY | hsa04540: Gap
junction | 8 | 0.046579 |
miRNA-gene pairs and miRNA-gene
network
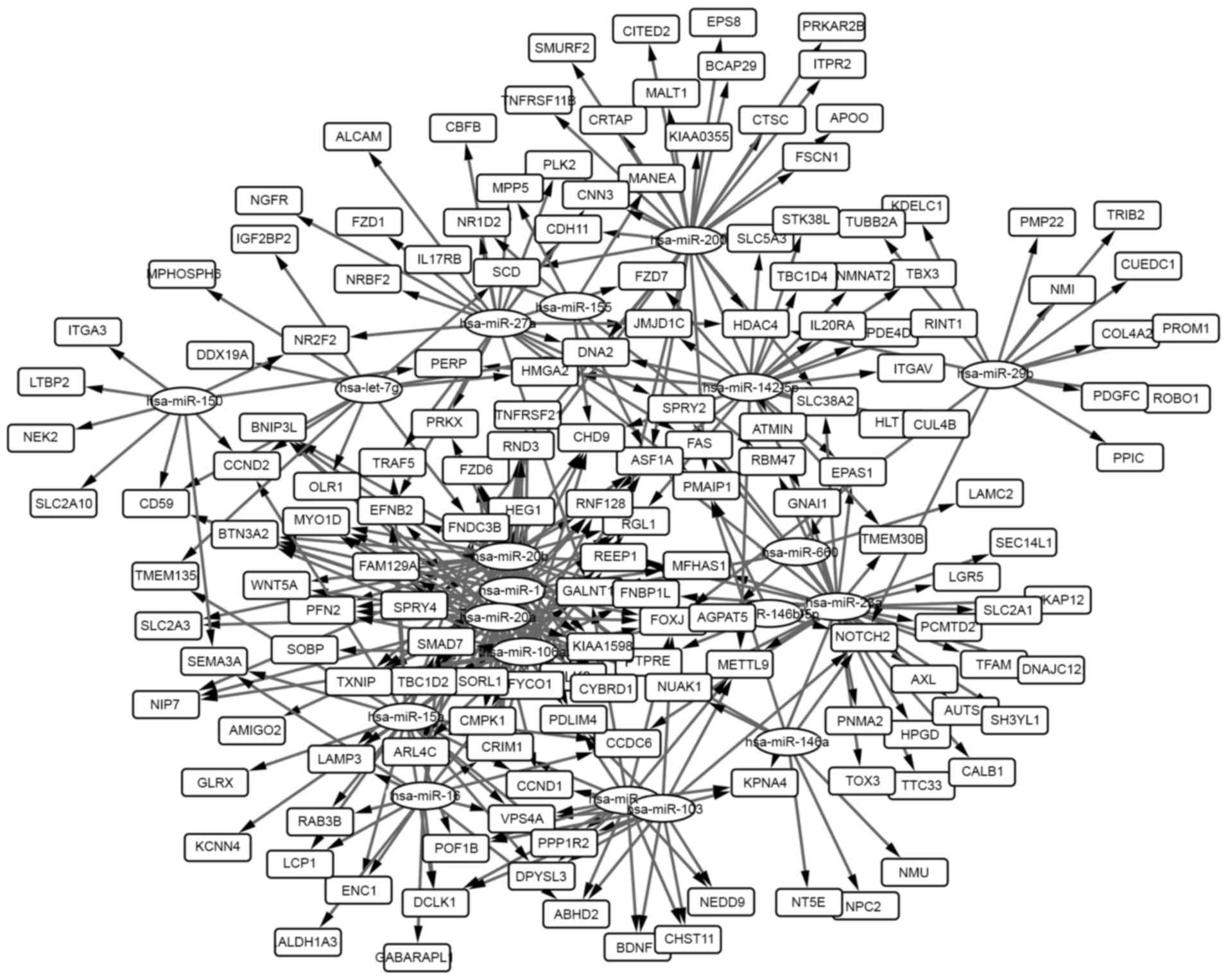
A total of 366 miRNA-gene pairs among the overlapped
genes with the DEMs were selected, and the miRNA-gene network was
generated and analyzed. The network is presented in Fig. 2 and the 20 highest degree nodes are
presented in Table V. The term
‘degree’ represented connections of one node with other nodes.
 | Table V.20 highest degree nodes in the
miRNA-gene network. |
Table V.
20 highest degree nodes in the
miRNA-gene network.
| Node | Degree |
|---|
|
hsa-miR-20b | 38 |
|
hsa-miR-20a | 36 |
|
hsa-miR-106a | 33 |
|
hsa-miR-23a | 32 |
|
hsa-miR-200b | 30 |
|
hsa-miR-17 | 26 |
|
hsa-miR-142-5p | 24 |
|
hsa-miR-15a | 23 |
|
hsa-miR-27a | 22 |
|
hsa-miR-107 | 17 |
|
hsa-miR-103 | 15 |
|
hsa-miR-16 | 15 |
|
hsa-miR-29b | 14 |
|
hsa-let-7g | 11 |
|
hsa-miR-150 | 9 |
| FNDC3B | 8 |
| CRIM1 | 7 |
| FOXJ2 | 7 |
|
hsa-miR-146a | 7 |
|
hsa-miR-155 | 7 |
Enriched pathways of predicted target
genes
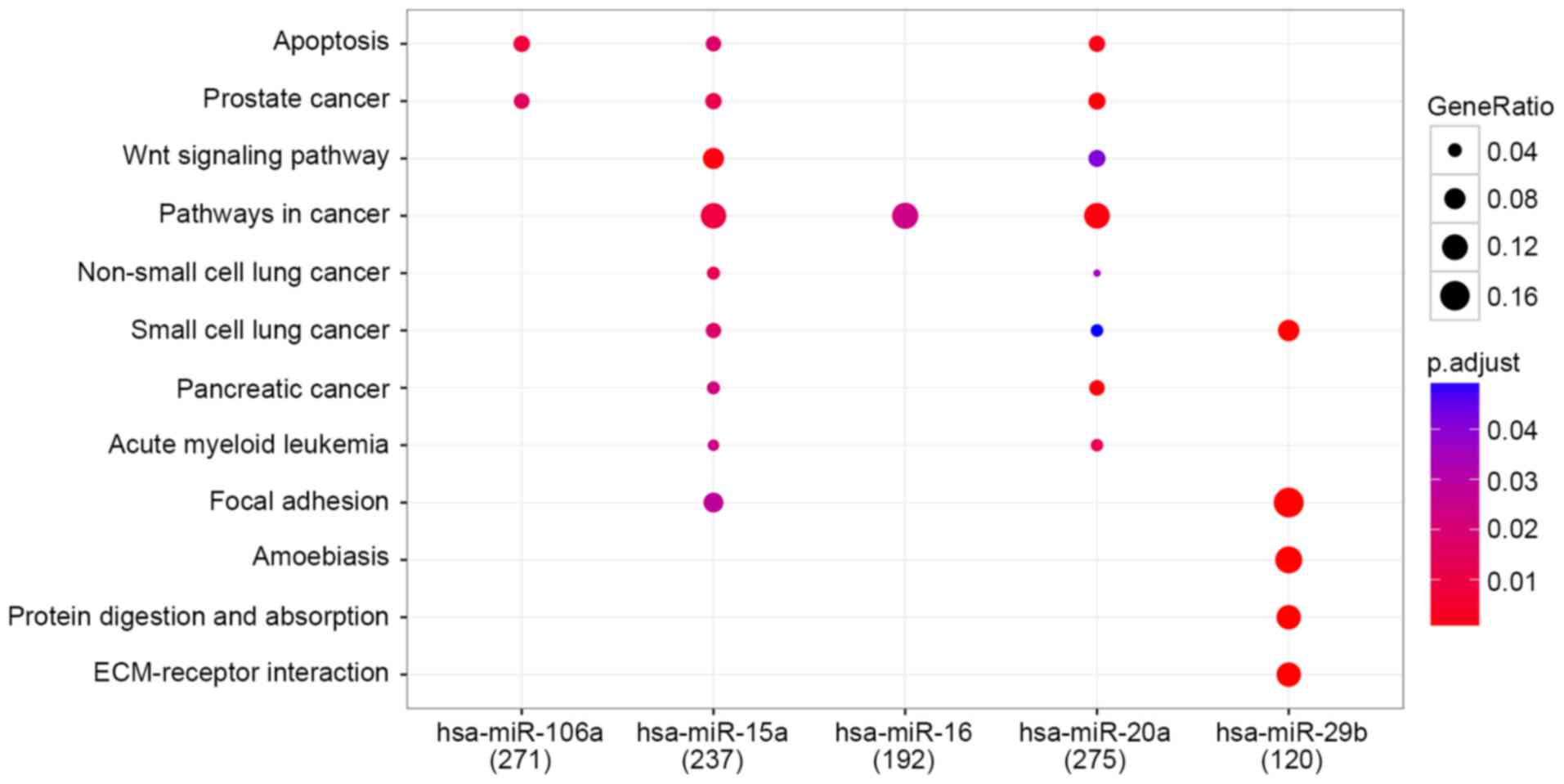
In total 271, 237, 192, 275 and 120 genes were
respectively regulated by each of the 5 miRNAs hsa-miR-106a,
hsa-miR-15a, hsa-miR-16, hsa-miR-20a and hsa-miR-29b,
respectively. Overall, 12 pathways were enriched and the results
are presented in Fig. 3.
Discussion
In the present study, DEGs and DEMs were initially
identified in colorectal cancer samples with lymph node metastasis
compared with primary colorectal cancer samples. Subsequently, the
functional and pathway enrichment analysis of DEGs and the
predicted target genes regulated by DEMs was performed. The
over-represented pathways were associated with pathways in cancer,
the Wnt signaling pathway and extracellular matrix (ECM)-receptor
interaction. The major enriched pathway of DEMs was pathways in
cancer characterized by the lowest P-values (Table IV). Overall, ~12% of predicted target
genes regulated by hsa-miR-15a, hsa-miR-16 and
hsa-miR-20a were associated with this pathway; suggesting
that it serves a critical role in CRC metastasis.
Wnt signaling is involved in embryonic development
(31) and a number of studies
demonstrated that aberrant Wnt signaling serves an important role
in CRC, regulating several cellular processes, including cell
migration and metastasis (32,33). Hu
et al (34) reported that
CXCR4 promotes CRC progression and epithelial-mesenchymal
transition by activating the Wnt/β-catenin signaling pathway. A
study by Ting et al (35)
indicated that the genetic interaction profile of Wnt pathway
genetic variants may increase the prognostic value of outcome
prediction for CRC patients. Therefore, it was indicated that the
Wnt signaling pathway may serve an important role in the processes
of cell migration and metastasis, and that certain genes in this
pathway may serve as potential metastatic biomarkers for CRC.
The ECM regulates tissue architecture and
adipogenesis, which involves a complex mixture of structural and
functional macromolecules, including glycosaminoglycans and fibrous
proteins (36). One recent study
revealed that twist-related protein 2 (Twist2) regulates the
expression of integrin α-4 and CD44 antigen, two major proteins in
the ECM-receptor interaction pathway (37). Furthermore, it was also demonstrated
that the overexpression of Twist2 may be involved in cell growth
regulation, apoptosis and motility, and that Twist2 may serve as a
potential therapeutic target for the treatment of kidney cancer
(37). Additionally, twist family
BHLH transcription factor 2 was significantly overexpressed in
several solid tumors and contributed to tumor progression (38). The results of the present study
suggest that ECM-receptor interaction may be associated with CRC
metastasis, however, further research is required to validate this
association.
miRNA-gene network analysis revealed that
fibronectin type III domain-containing 3B (FNDC3B), cysteine
rich transmembrane BMP regulator 1 (CRIM1) and forkhead box
J2 (FOXJ2) were the genes with the highest degree (Table V). It has been demonstrated that
FNDC3B is a positive regulator of adipocyte differentiation,
and that it suppresses the invasion and metastasis of melanoma
cells (39). FNDC3B mutations
were associated with rapid postnatal death and the inhibition of
cellular proliferation, adhesion and migration (40). FNDC3B has been associated with
the activation of several cancer pathways and tumor progression
(41). CRIM1 encodes the
cysteine-rich motor neuron 1 protein (CRIM1), which has been
characterized as a potential cancer biomarker (42). It has been reported that increased
CRIM1 inhibits the proliferation and migration of vascular
endothelial cells (43).
Additionally, increased CRIM1 expression has been reported
in drug-resistant myeloid leukemia cells compared with
drug-sensitive cells (44).
FOXJ2 serves an important role in the migration of glioma
cells (45) and FOXJ2
overexpression decreases the migration of breast cancer cells
(46). Furthermore, abnormal
expression of FOXJ2 suppressed migration and invasion in
extrahepatic cholangiocarcinoma, which was associated with an
improved prognosis (47).
FNDC3B, CRIM1 and FOXJ2 have been associated
with tumor migration and prognosis. The findings of the present
study suggest that they may also be associated with CRC
metastasis.
In conclusion, the present study demonstrated that
DEGs and predicted target genes of the DEMs are enriched in
pathways in cancer, the Wnt signaling pathway and ECM-receptor
interaction, which may serve a critical role in the metastatic
mechanism of CRC. Furthermore, FNDC3B, CRIM1 and
FOXJ2 are proposed as potential biomarkers for metastatic
CRC.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar
|
|
2
|
Shike M, Winawer SJ, Greenwald PH, Bloch
A, Hill MJ and Swaroop SV: Primary prevention of colorectal cancer.
The WHO collaborating centre for the prevention of colorectal
cancer. Bull World Health Organ. 68:377–385. 1990.
|
|
3
|
Matsuda T, Ono A, Kakugawa Y, Matsumoto M
and Saito Y: Impact of screening colonoscopy on outcomes in
colorectal cancer. Jpn J Clin Oncol. 45:900–905. 2015. View Article : Google Scholar
|
|
4
|
Sabatino SA, Lawrence B, Elder R, Mercer
SL, Wilson KM, DeVinney B, Melillo S, Carvalho M, Taplin S, Bastani
R, et al: Effectiveness of interventions to increase screening for
breast, cervical and colorectal cancers: Nine updated systematic
reviews for the guide to community preventive services. Am J Prev
Med. 43:97–118. 2012. View Article : Google Scholar
|
|
5
|
Board PDQATE: Colon Cancer Treatment
(PDQ®): Health professional versionPDQ Cancer
Information Summaries. National Cancer Institute; Bethesda, MD:
2002
|
|
6
|
Cao H, Xu E, Liu H, Wan L and Lai M:
Epithelial-mesenchymal transition in colorectal cancer metastasis:
A system review. Pathol Res Pract. 211:557–569. 2015. View Article : Google Scholar
|
|
7
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar
|
|
8
|
Christofori G: New signals from the
invasive front. Nature. 441:444–450. 2006. View Article : Google Scholar
|
|
9
|
Kanthan R, Senger JL and Kanthan SC:
Molecular events in primary and metastatic colorectal carcinoma: A
review. Patholog Res Int. 2012:5974972012.
|
|
10
|
Siegel R, DeSantis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar
|
|
11
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
12
|
Grothey A and Schmoll HJ: New chemotherapy
approaches in colorectal cancer. Curr Opin Oncol. 13:275–286. 2001.
View Article : Google Scholar
|
|
13
|
Fidler IJ: The pathogenesis of cancer
metastasis: The ‘seed and soil’ hypothesis revisited. Nat Rev
Cancer. 3:453–458. 2003. View
Article : Google Scholar
|
|
14
|
Cui YM, Jiao HL, Ye YP, Chen CM, Wang JX,
Tang N, Li TT, Lin J, Qi L, Wu P, et al: FOXC2 promotes colorectal
cancer metastasis by directly targeting MET. Oncogene.
34:4379–4390. 2015. View Article : Google Scholar
|
|
15
|
Yuan R, Ke J, Sun L, He Z, Zou Y, He X,
Chen Y, Wu X, Cai Z, Wang L, et al: HES1 promotes metastasis and
predicts poor survival in patients with colorectal cancer. Clin Exp
Metastasis. 32:169–179. 2015. View Article : Google Scholar
|
|
16
|
Zhang Z, Liu X, Feng B, Liu N, Wu Q, Han
Y, Nie Y, Wu K, Shi Y and Fan D: STIM1, a direct target of
microRNA-185, promotes tumor metastasis and is associated with poor
prognosis in colorectal cancer. Oncogene. 34:4808–4820. 2015.
View Article : Google Scholar
|
|
17
|
Zhou ZH, Rao J, Yang J, Wu F, Tan J, Xu
SL, Ding Y, Zhan N, Hu XG, Cui YH, et al: SEMA3F prevents
metastasis of colorectal cancer by PI3K-AKT-dependent
down-regulation of the ASCL2-CXCR4 axis. J Pathol. 236:467–478.
2015. View Article : Google Scholar
|
|
18
|
Fang Y, Liang X, Jiang W, Li J, Xu J and
Cai X: Cyclin b1 suppresses colorectal cancer invasion and
metastasis by regulating e-cadherin. PLoS One. 10:e01268752015.
View Article : Google Scholar
|
|
19
|
Li X, Chen T, Shi Q, Li J, Cai S, Zhou P,
Zhong Y and Yao L: Angiopoietin-like 4 enhances metastasis and
inhibits apoptosis via inducing bone morphogenetic protein 7 in
colorectal cancer cells. Biochem Biophys Res Commun. 467:128–134.
2015. View Article : Google Scholar
|
|
20
|
Song B, Wang W, Zheng Y, Yang J and Xu Z:
P21-activated kinase 1 and 4 were associated with colorectal cancer
metastasis and infiltration. J Surg Res. 196:130–135. 2015.
View Article : Google Scholar
|
|
21
|
Purnak T, Ozaslan E and Efe C: Molecular
basis of colorectal cancer. N Engl J Med. 362:1246–1247. 2010.
|
|
22
|
Provenzani A, Fronza R, Loreni F, Pascale
A, Amadio M and Quattrone A: Global alterations in mRNA polysomal
recruitment in a cell model of colorectal cancer progression to
metastasis. Carcinogenesis. 27:1323–1333. 2006. View Article : Google Scholar
|
|
23
|
Drusco A, Nuovo GJ, Zanesi N, Di Leva G,
Pichiorri F, Volinia S, Fernandez C, Antenucci A, Costinean S,
Bottoni A, et al: MicroRNA profiles discriminate among colon cancer
metastasis. PLoS One. 9:e966702014. View Article : Google Scholar
|
|
24
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar
|
|
25
|
Wiberg AO, Liu L, Tong Z, Myslivets E,
Ataie V, Kuo BP, Alic N and Radic S: Photonic preprocessor for
analog-to-digital-converter using a cavity-less pulse source. Opt
Express. 20:B419–B427. 2012. View Article : Google Scholar
|
|
26
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar
|
|
27
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar
|
|
28
|
Dweep H, Gretz N and Sticht C: miRWalk
database for miRNA-target interactions. Methods Mol Biol.
1182:289–305. 2014. View Article : Google Scholar
|
|
29
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar
|
|
30
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar
|
|
31
|
Heuberger J and Birchmeier W: Interplay of
cadherin-mediated cell adhesion and canonical Wnt signaling. Cold
Spring Harb Perspect Biol. 2:a0029152010. View Article : Google Scholar
|
|
32
|
Tenbaum SP, Ordóñez-Morán P, Puig I,
Chicote I, Arqués O, Landolfi S, Fernández Y, Herance JR, Gispert
JD, Mendizabal L, et al: β-catenin confers resistance to PI3K and
AKT inhibitors and subverts FOXO3a to promote metastasis in colon
cancer. Nat Med. 18:892–901. 2012. View
Article : Google Scholar
|
|
33
|
Qi J, Yu Y, Öztürk Ö Akilli, Holland JD,
Besser D, Fritzmann J, Wulf-Goldenberg A, Eckert K, Fichtner I and
Birchmeier W: New Wnt/β-catenin target genes promote experimental
metastasis and migration of colorectal cancer cells through
different signals. Gut. 65:1690–1701. 2016. View Article : Google Scholar
|
|
34
|
Hu TH, Yao Y, Yu S, Han LL, Wang WJ, Guo
H, Tian T, Ruan ZP, Kang XM, Wang J, et al: SDF-1/CXCR4 promotes
epithelial-mesenchymal transition and progression of colorectal
cancer by activation of the Wnt/β-catenin signaling pathway. Cancer
Lett. 354:417–426. 2014. View Article : Google Scholar
|
|
35
|
Ting WC, Chen LM, Pao JB, Yang YP, You BJ,
Chang TY, Lan YH, Lee HZ and Bao BY: Common genetic variants in Wnt
signaling pathway genes as potential prognostic biomarkers for
colorectal cancer. PLoS One. 8:e561962013. View Article : Google Scholar
|
|
36
|
Mariman EC and Wang P: Adipocyte
extracellular matrix composition, dynamics and role in obesity.
Cell Mol Life Sci. 67:1277–1292. 2010. View Article : Google Scholar
|
|
37
|
Zhang HJ, Tao J, Sheng L, Hu X, Rong RM,
Xu M and Zhu TY: Twist2 promotes kidney cancer cell proliferation
and invasion by regulating ITGA6 and CD44 expression in the
ECM-receptor interaction pathway. Onco Targets Ther. 9:1801–1812.
2016.
|
|
38
|
Ansieau S, Bastid J, Doreau A, Morel AP,
Bouchet BP, Thomas C, Fauvet F, Puisieux I, Doglioni C, Piccinin S,
et al: Induction of EMT by twist proteins as a collateral effect of
tumor-promoting inactivation of premature senescence. Cancer Cell.
14:79–89. 2008. View Article : Google Scholar
|
|
39
|
Katoh D, Nishizuka M, Osada S and Imagawa
M: Fad104, a positive regulator of adipocyte differentiation,
suppresses invasion and metastasis of melanoma cells by inhibition
of STAT3 activity. PLoS One. 10:e01171972015. View Article : Google Scholar
|
|
40
|
Nishizuka M, Kishimoto K, Kato A, Ikawa M,
Okabe M, Sato R, Niida H, Nakanishi M, Osada S and Imagawa M:
Disruption of the novel gene fad104 causes rapid postnatal death
and attenuation of cell proliferation, adhesion, spreading and
migration. Exp Cell Res. 315:809–819. 2009. View Article : Google Scholar
|
|
41
|
Cai C, Rajaram M, Zhou X, Liu Q, Marchica
J, Li J and Powers RS: Activation of multiple cancer pathways and
tumor maintenance function of the 3q amplified oncogene FNDC3B.
Cell Cycle. 11:1773–1781. 2012. View
Article : Google Scholar
|
|
42
|
Zeng H and Tang L: CRIM1, the antagonist
of BMPs, is a potential risk factor of cancer. Curr Cancer Drug
Targets. 14:652–658. 2014. View Article : Google Scholar
|
|
43
|
Nakashima Y, Morimoto M, Toda K, Shinya T,
Sato K and Takahashi S: Inhibition of the proliferation and
acceleration of migration of vascular endothelial cells by
increased cysteine-rich motor neuron 1. Biochem Biophys Res Commun.
462:215–220. 2015. View Article : Google Scholar
|
|
44
|
Prenkert M, Uggla B, Tidefelt U and Strid
H: CRIM1 is expressed at higher levels in drug-resistant than in
drug-sensitive myeloid leukemia HL60 cells. Anticancer Res.
30:4157–4161. 2010.
|
|
45
|
Qiu X, Ji B, Yang L, Huang Q, Shi W, Ding
Z, He X, Ban N, Fan S, Zhang J and Tian Y: The role of FoxJ2 in the
migration of human glioma cells. Pathol Res Pract. 211:389–397.
2015. View Article : Google Scholar
|
|
46
|
Wang Y, Yang S, Ni Q, He S, Zhao Y, Yuan
Q, Li C, Chen H, Zhang L, Zou L, et al: Overexpression of forkhead
box J2 can decrease the migration of breast cancer cells. J Cell
Biochem. 113:2729–2737. 2012. View Article : Google Scholar
|
|
47
|
Qiang Y, Wang F, Yan S, Zhang H, Zhu L and
Chen Z, Tu F, Wang D, Wang G, Wang W and Chen Z: Abnormal
expression of Forkhead Box J2 (FOXJ2) suppresses migration and
invasion in extrahepatic cholangiocarcinoma and is associated with
prognosis. Int J Oncol. 46:2449–2458. 2015. View Article : Google Scholar
|