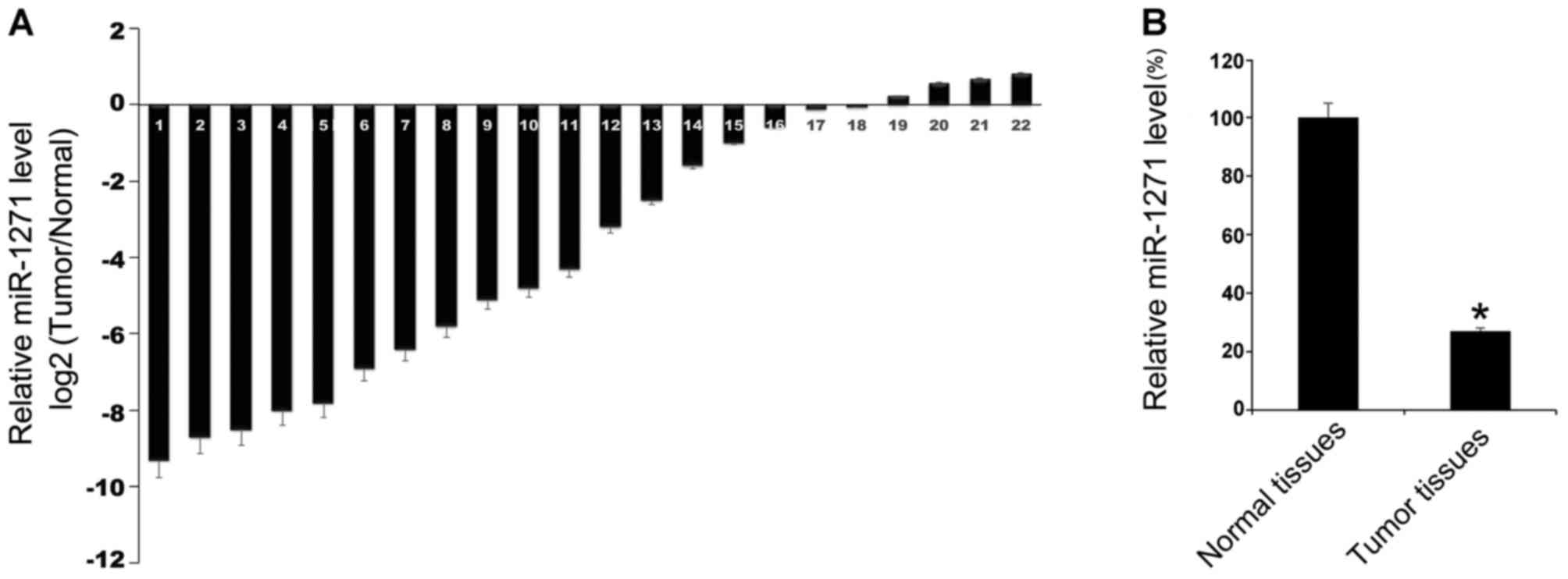
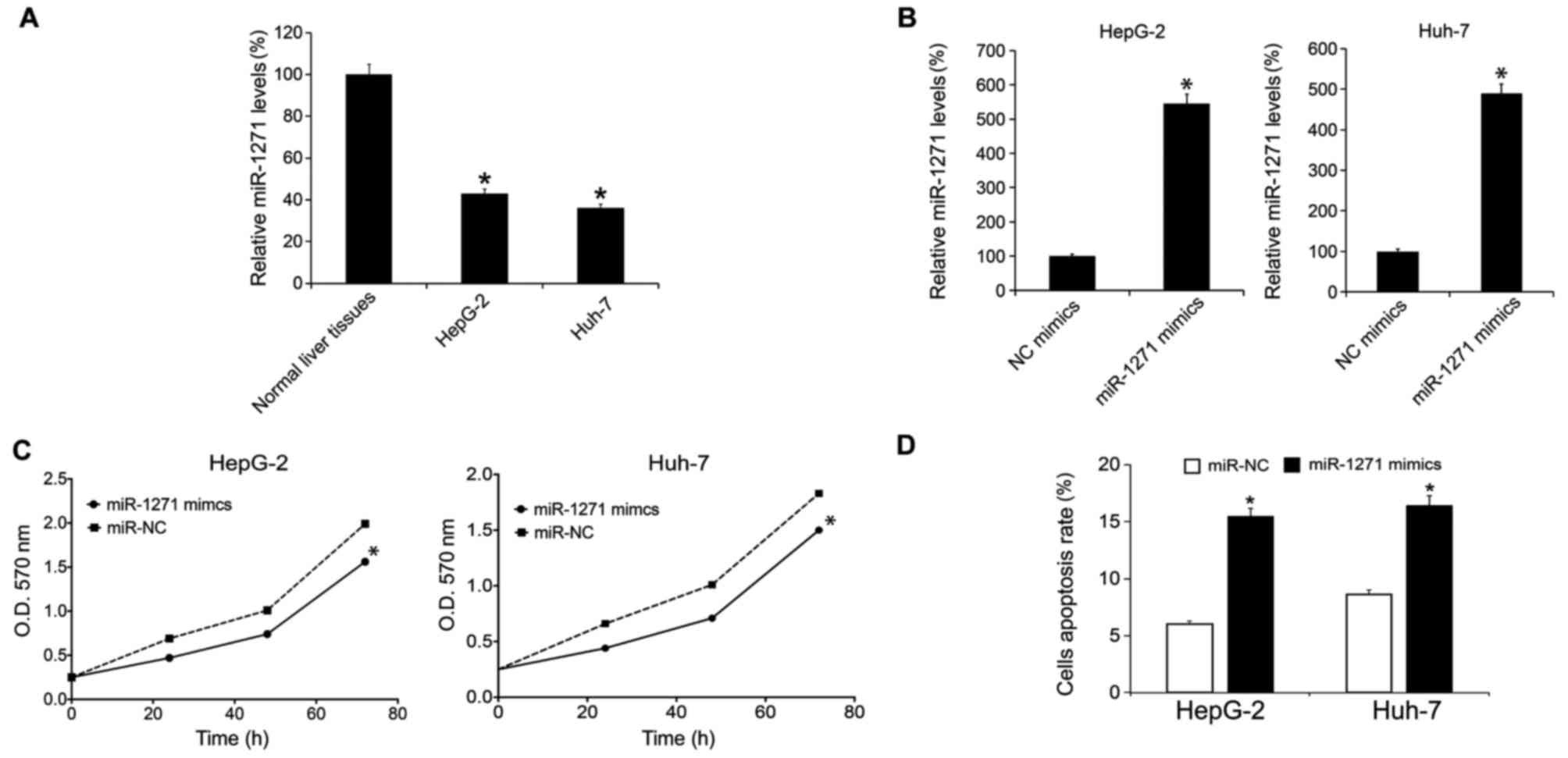
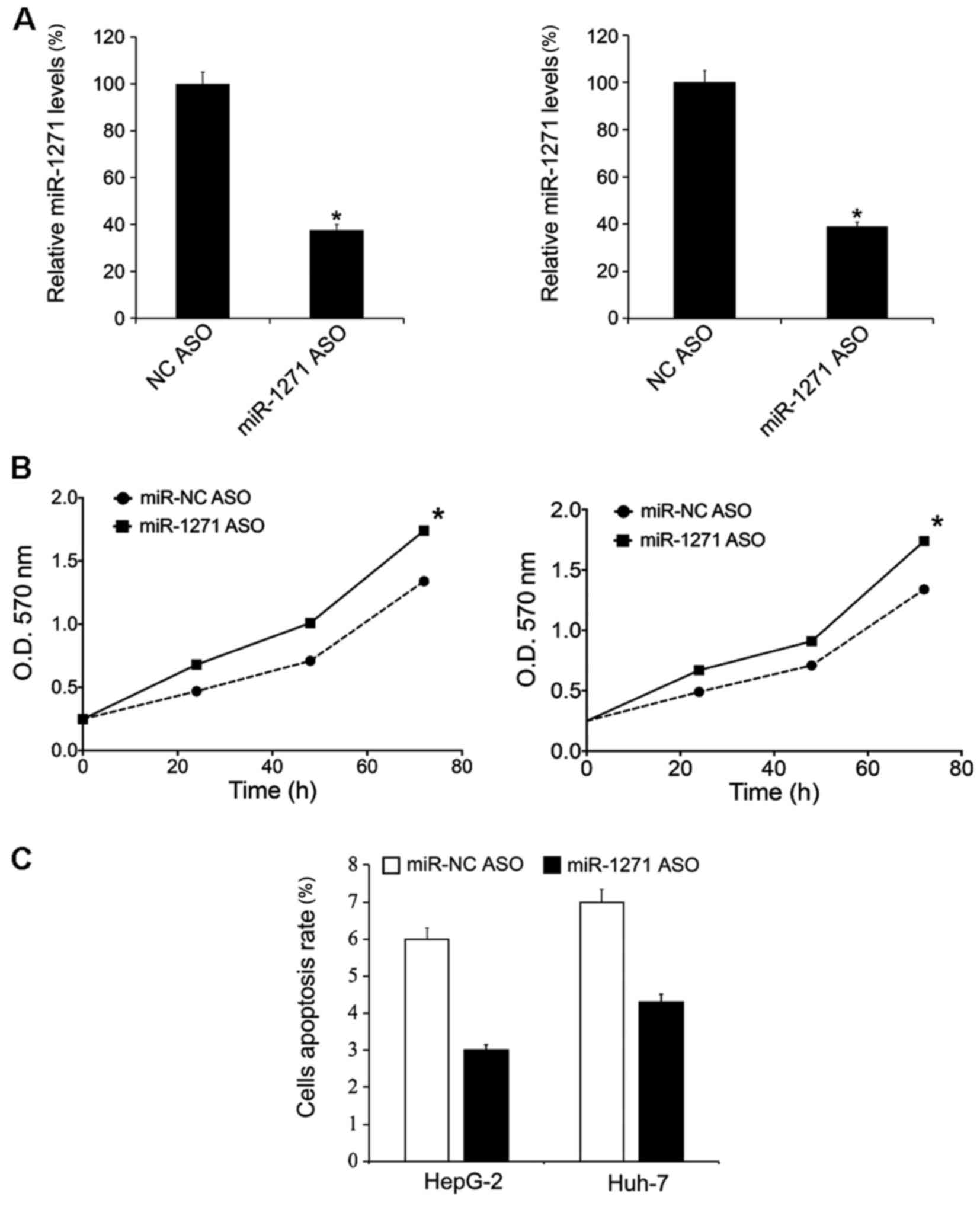
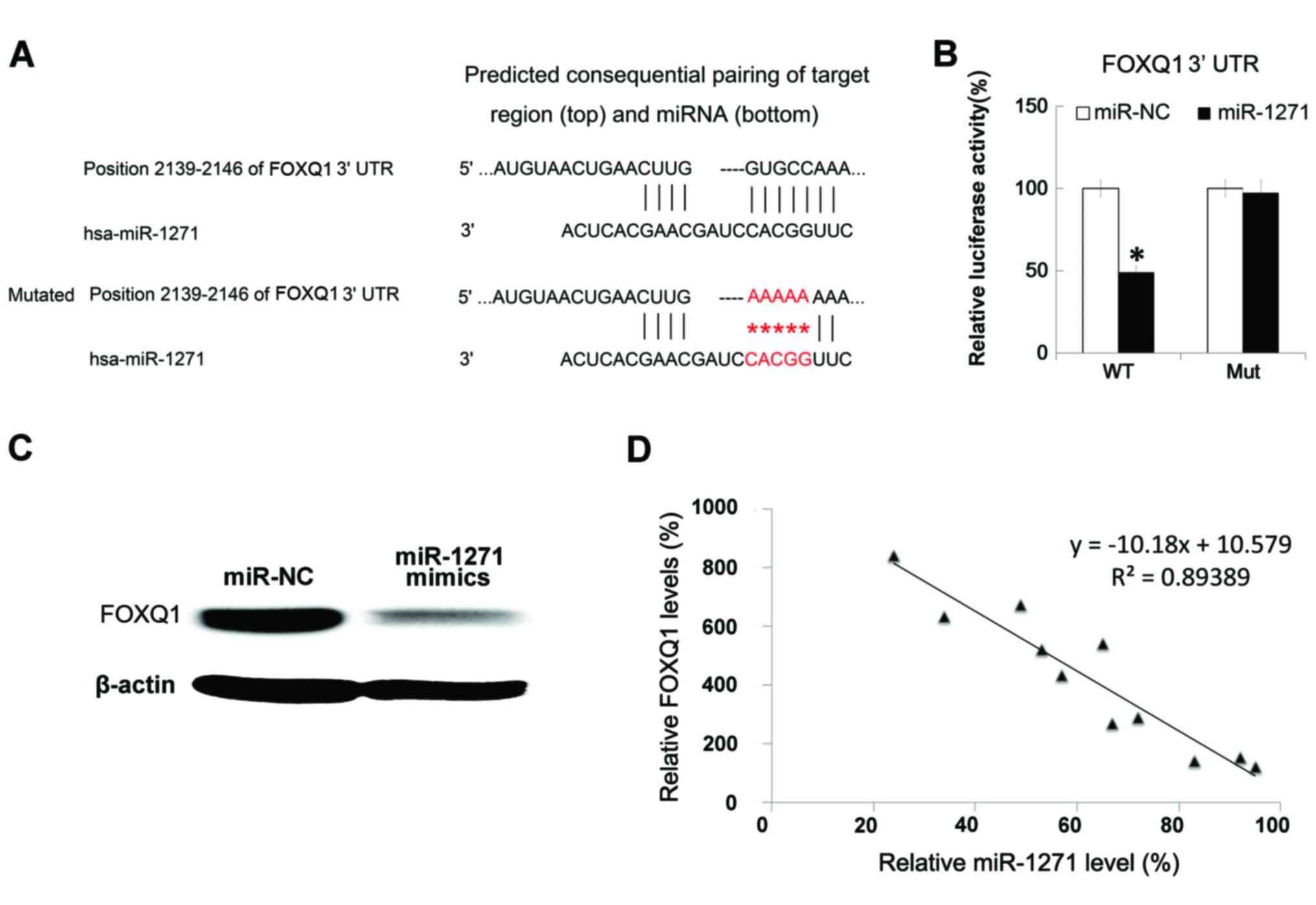
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Venook AP, Papandreou C, Furuse J and de
Guevara LL: The incidence and epidemiology of hepatocellular
carcinoma: A global and regional perspective. Oncologist. 15 Suppl
4:S5–S13. 2010. View Article : Google Scholar
|
|
4
|
Beasley RP: Hepatitis B virus. The major
etiology of hepatocellular carcinoma. Cancer. 61:1942–1956. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Song P, Feng X, Zhang K, Song T, Ma K,
Kokudo N, Dong J, Yao L and Tang W: Screening for and surveillance
of high-risk patients with HBV-related chronic liver disease:
Promoting the early detection of hepatocellular carcinoma in China.
Biosci Trends. 7:1–6. 2013.PubMed/NCBI
|
|
6
|
Karaman B, Battal B, Sari S and Verim S:
Hepatocellular carcinoma review: Current treatment, and
evidence-based medicine. World J Gastroenterol. 20:18059–18060.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gramantieri L, Ferracin M, Fornari F,
Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E,
Grazi GL, et al: Cyclin G1 is a target of miR-122a, a microRNA
frequently down-regulated in human hepatocellular carcinoma. Cancer
Res. 67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Budhu A, Jia HL, Forgues M, Liu CG,
Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al:
Identification of metastasis-related microRNAs in hepatocellular
carcinoma. Hepatology. 47:897–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP and
Zhuang SM: MicroRNA-195 suppresses tumorigenicity and regulates
G1/S transition of human hepatocellular carcinoma cells.
Hepatology. 50:113–121. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y
and Zhuang SM: MicroRNA-101, down-regulated in hepatocellular
carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer
Res. 69:1135–1142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Murakami Y, Yasuda T, Saigo K, Urashima T,
Toyoda H, Okanoue T and Shimotohno K: Comprehensive analysis of
microRNA expression patterns in hepatocellular carcinoma and
non-tumorous tissues. Oncogene. 25:2537–2545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Braconi C and Patel T: MicroRNA expression
profiling: A molecular tool for defining the phenotype of
hepatocellular tumors. Hepatology. 47:1807–1809. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Murakami Y, Tamori A, Itami S, Tanahashi
T, Toyoda H, Tanaka M, Wu W, Brojigin N, Kaneoka Y, Maeda A, et al:
The expression level of miR-18b in hepatocellular carcinoma is
associated with the grade of malignancy and prognosis. BMC Cancer.
13:992013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gramantieri L, Fornari F, Callegari E,
Sabbioni S, Lanza G, Croce CM, Bolondi L and Negrini M: MicroRNA
involvement in hepatocellular carcinoma. J Cell Mol Med.
12:2189–2204. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang B, Majumder S, Nuovo G, Kutay H,
Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K and Jacob ST:
Role of microRNA-155 at early stages of hepatocarcinogenesis
induced by choline-deficient and amino acid-defined diet in C57BL/6
mice. Hepatology. 50:1152–1161. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Coulouarn C, Factor VM, Andersen JB,
Durkin ME and Thorgeirsson SS: Loss of miR-122 expression in liver
cancer correlates with suppression of the hepatic phenotype and
gain of metastatic properties. Oncogene. 28:3526–3536. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ji J, Shi J, Budhu A, Yu Z, Forgues M,
Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ji J, Yamashita T, Budhu A, Forgues M, Jia
HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH, et al: Identification
of microRNA-181 by genome-wide screening as a critical player in
EpCAM-positive hepatic cancer stem cells. Hepatology. 50:472–480.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ladeiro Y, Couchy G, Balabaud C,
Bioulac-Sage P, Pelletier L, Rebouissou S and Zucman-Rossi J:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song G, Sharma AD, Roll GR, Ng R, Lee AY,
Blelloch RH, Frandsen NM and Willenbring H: MicroRNAs control
hepatocyte proliferation during liver regeneration. Hepatology.
51:1735–1743. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ura S, Honda M, Yamashita T, Ueda T,
Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K and Kaneko
S: Differential microRNA expression between hepatitis B and
hepatitis C leading disease progression to hepatocellular
carcinoma. Hepatology. 49:1098–1112. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as potential oncogenic microRNA of
squamous cell carcinoma of tongue. Clin Cancer Res. 14:2588–2592.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Y, Lee AT, Ma JZ, Wang J, Ren J, Yang
Y, Tantoso E, Li KB, Ooi LL, Tan P and Lee CG: Profiling microRNA
expression in hepatocellular carcinoma reveals microRNA-224
up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific
target. J Biol Chem. 283:13205–13215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiang XJ, Deng J, Liu YW, Wan LY, Feng M,
Chen J and Xiong JP: MiR-1271 inhibits cell proliferation, invasion
and EMT in gastric cancer by targeting FOXQ1. Cell Physiol Biochem.
36:1382–1394. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kong D, Zhang G, Ma H and Jiang G:
miR-1271 inhibits OSCC cell growth and metastasis by targeting ALK.
Neoplasma. 62:559–566. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maurel M, Jalvy S, Ladeiro Y, Combe C,
Vachet L, Sagliocco F, Bioulac-Sage P, Pitard V, Jacquemin-Sablon
H, Zucman-Rossi J, et al: A functional screening identifies five
microRNAs controlling glypican-3: Role of miR-1271 down-regulation
in hepatocellular carcinoma. Hepatology. 57:195–204. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fischer AH, Jacobson KA, Rose J and Zeller
R: Hematoxylin and eosin staining of tissue and cell sections. CSH
Protoc. 2008:pdb.prot49862008.PubMed/NCBI
|
|
31
|
Li D, Liu X, Lin L, Hou J, Li N, Wang C,
Wang P, Zhang Q, Zhang P, Zhou W, et al: MicroRNA-99a inhibits
hepatocellular carcinoma growth and correlates with prognosis of
patients with hepatocellular carcinoma. J Biol Chem.
286:36677–36685. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Song B, Zhang C, Li G, Jin G and Liu C:
MiR-940 inhibited pancreatic ductal adenocarcinoma growth by
targeting MyD88. Cell Physiol Biochem. 35:1167–1177. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mosmann T: Rapid colorimetric assay for
cellular growth and survival: Application to proliferation and
cytotoxicity assays. J Immunol Methods. 65:55–63. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: Determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Garcia DM, Baek D, Shin C, Bell GW,
Grimson A and Bartel DP: Weak seed-pairing stability and high
target-site abundance decrease the proficiency of lsy-6 and other
microRNAs. Nat Struct Mol Biol. 18:1139–1146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Grentzmann G, Ingram JA, Kelly PJ,
Gesteland RF and Atkins JF: A dual-luciferase reporter system for
studying recoding signals. RNA. 4:479–486. 1998.PubMed/NCBI
|
|
40
|
Zhang H, Cai X, Wang Y, Tang H, Tong D and
Ji F: microRNA-143, down-regulated in osteosarcoma, promotes
apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol
Rep. 24:1363–1369. 2010.PubMed/NCBI
|
|
41
|
Carlsson P and Mahlapuu M: Forkhead
transcription factors: Key players in development and metabolism.
Dev Biol. 250:1–23. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Abba M, Patil N, Rasheed K, Nelson LD,
Mudduluru G, Leupold JH and Allgayer H: Unraveling the role of
FOXQ1 in colorectal cancer metastasis. Mol Cancer Res.
11:1017–1028. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qiao Y, Jiang X, Lee ST, Karuturi RK, Hooi
SC and Yu Q: FOXQ1 regulates epithelial-mesenchymal transition in
human cancers. Cancer Res. 71:3076–3086. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kaneda H, Arao T, Tanaka K, Tamura D,
Aomatsu K, Kudo K, Sakai K, De Velasco MA, Matsumoto K, Fujita Y,
et al: FOXQ1 is overexpressed in colorectal cancer and enhances
tumorigenicity and tumor growth. Cancer Res. 70:2053–2063. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Feng J, Zhang X, Zhu H, Wang X, Ni S and
Huang J: FoxQ1 overexpression influences poor prognosis in
non-small cell lung cancer, associates with the phenomenon of EMT.
PLoS One. 7:e399372012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang W, He S, Ji J, Huang J, Zhang S and
Zhang Y: The prognostic significance of FOXQ1 oncogene
overexpression in human hepatocellular carcinoma. Pathol Res Pract.
209:353–358. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xia L, Huang W, Tian D, Zhang L, Qi X,
Chen Z, Shang X, Nie Y and Wu K: Forkhead box Q1 promotes
hepatocellular carcinoma metastasis by transactivating ZEB2 and
VersicanV1 expression. Hepatology. 59:958–973. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fan DM, Feng XS, Qi PW and Chen YW:
Forkhead factor FOXQ1 promotes TGF-b1 expression and induces
epithelial-mesenchymal transition. Mol Cell Biochem. 397:179–186.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Christensen J, Bentz S, Sengstag T,
Shastri VP and Anderle P: FOXQ1, a novel target of the Wnt pathway
and a new marker for activation of Wnt signaling in solid tumors.
PLoS One. 8:e600512013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Peng XH, Huang HR, Lu J, Liu X, Zhao FP,
Zhang B, Lin SX, Wang L, Chen HH, Xu X, et al: MiR-124 suppresses
tumor growth and metastasis by targeting Foxq1 in nasopharyngeal
carcinoma. Mol Cancer. 13:1862014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang J, Yang Y, Yang T, Yuan S, Wang R,
Pan Z, Yang Y, Huang G, Gu F, Jiang B, et al: Double-negative
feedback loop between microRNA-422a and forkhead box (FOX)G1/Q1/E1
regulates hepatocellular carcinoma tumor growth and metastasis.
Hepatology. 61:561–573. 2015. View Article : Google Scholar : PubMed/NCBI
|