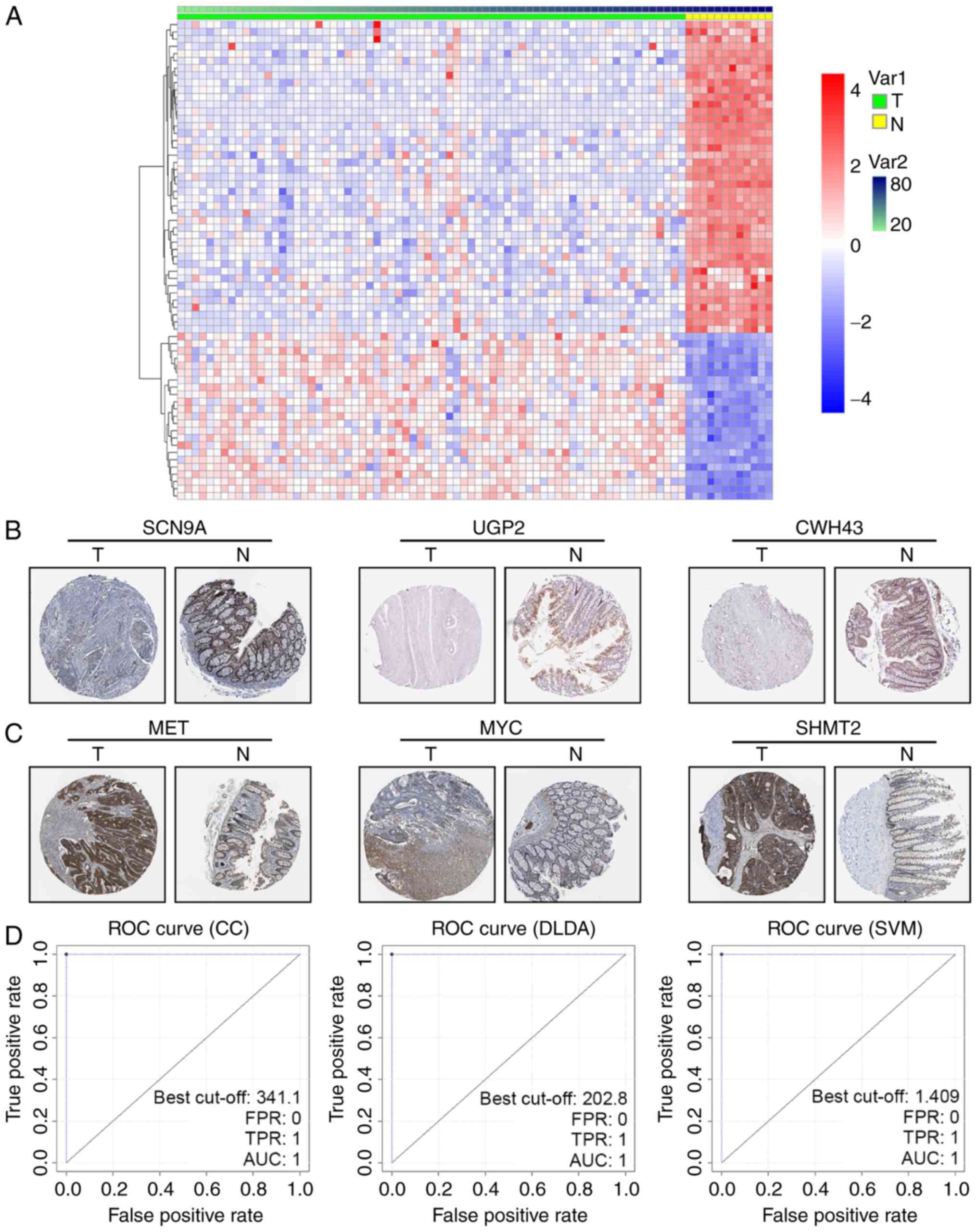
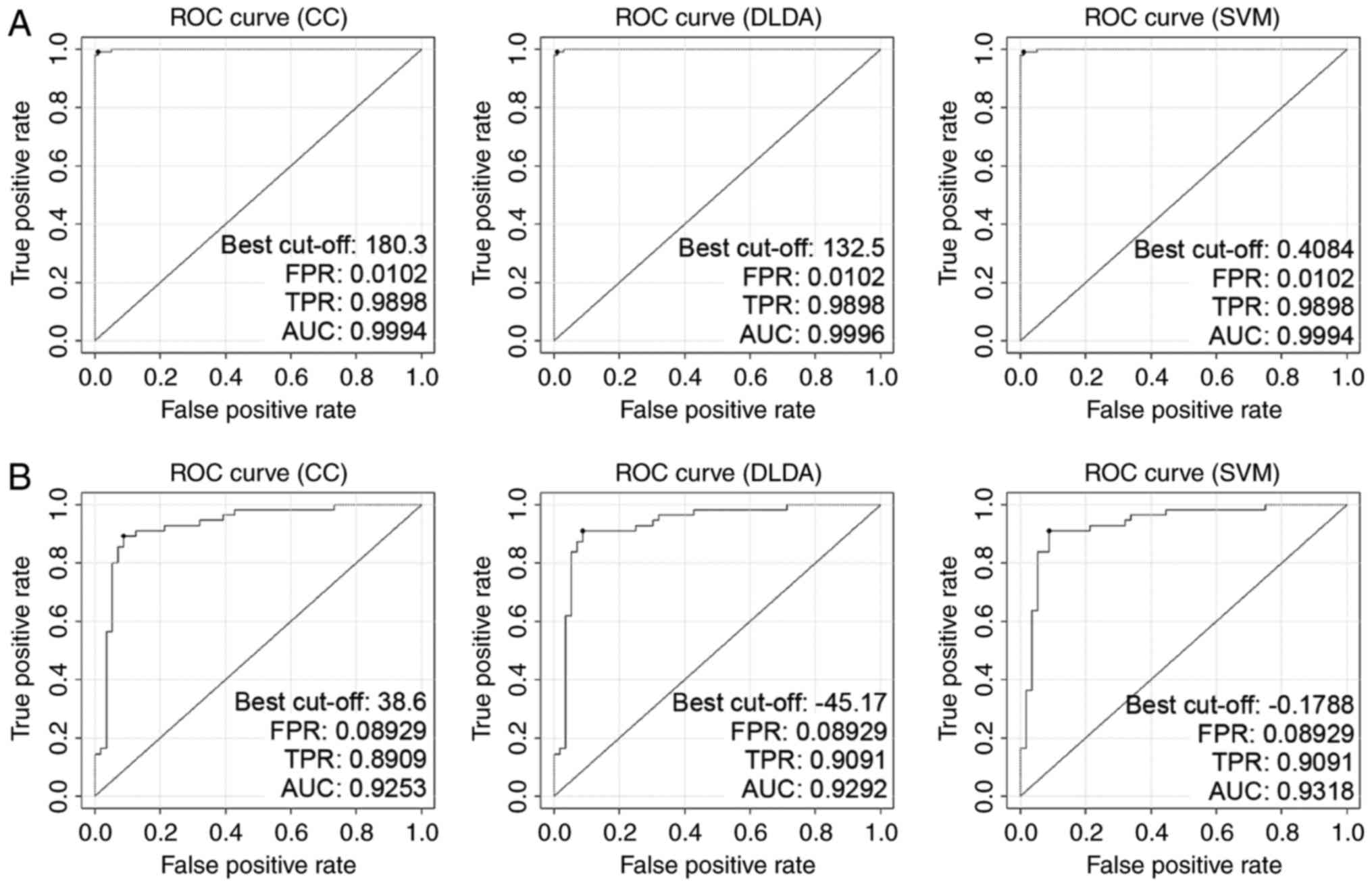
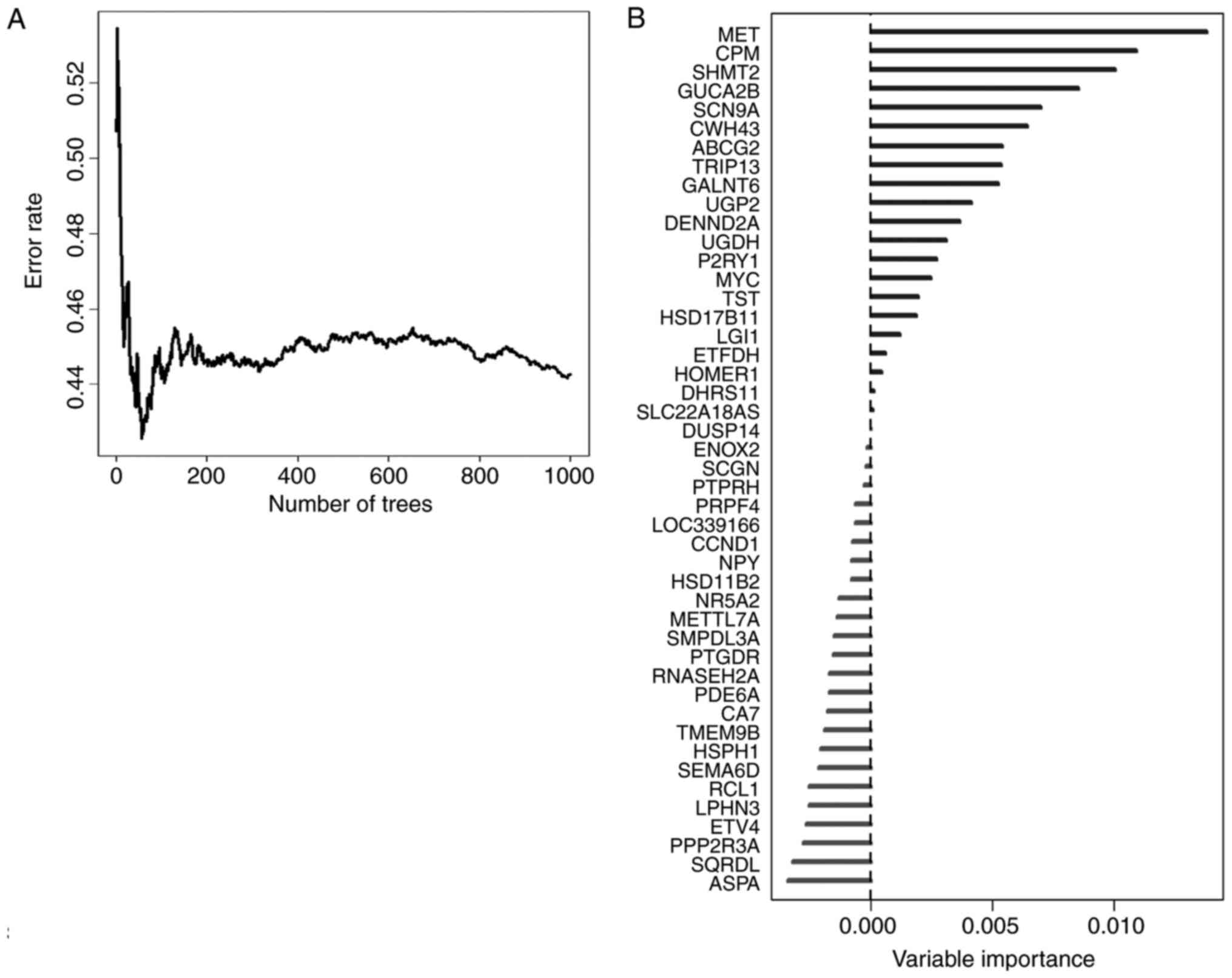
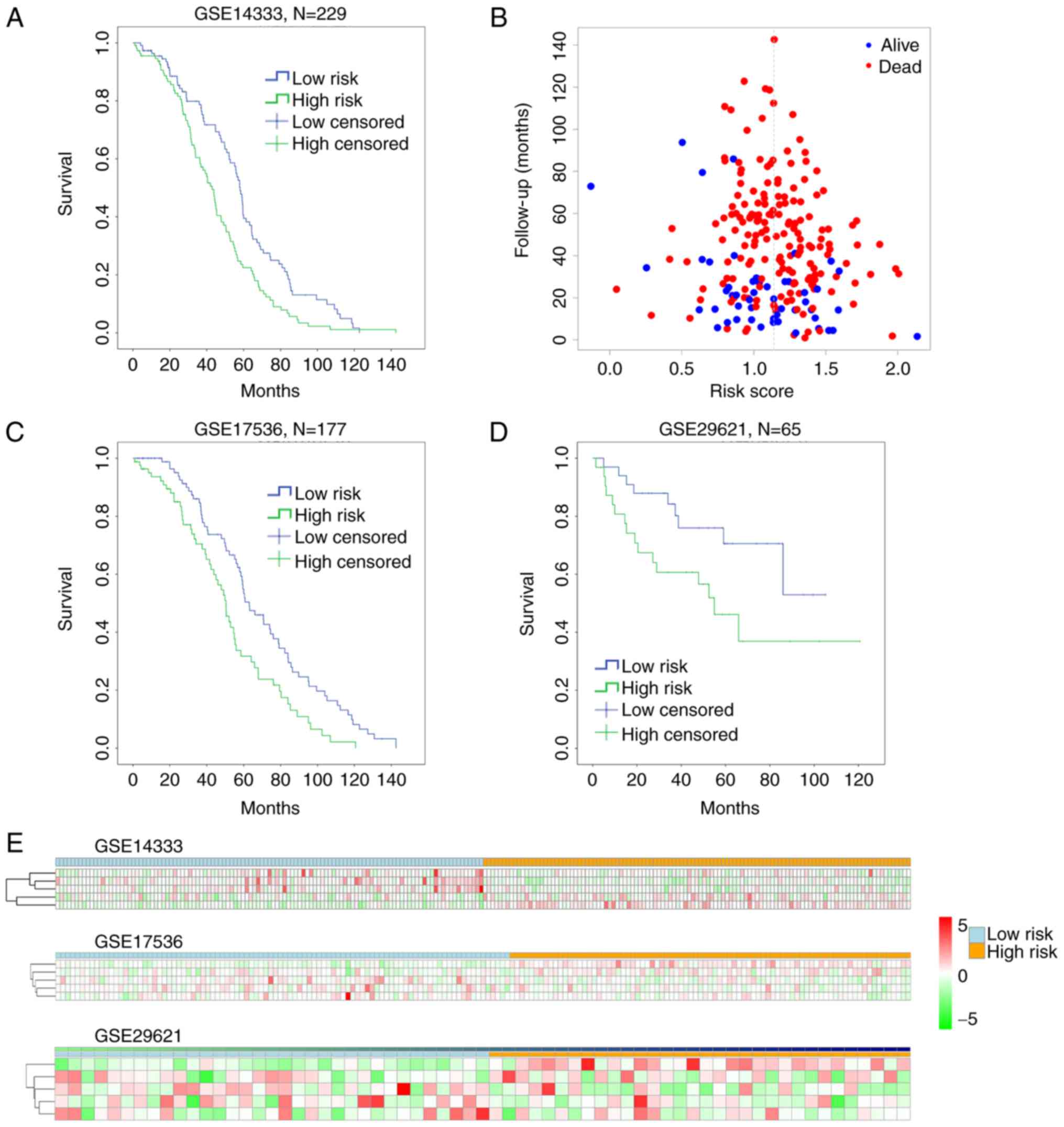
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Frampton M and Houlston RS: Modeling the
prevention of colorectal cancer from the combined impact of host
and behavioral risk factors. Genet Med. 19:314–321. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Inadomi JM: Screening for colorectal
neoplasia. N Engl J Med. 376:149–156. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chang W, Gao X, Han Y, Du Y, Liu Q, Wang
L, Tan X, Zhang Q, Liu Y, Zhu Y, et al: Gene expression
profiling-derived immunohistochemistry signature with high
prognostic value in colorectal carcinoma. Gut. 63:1457–1467. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shahid M, Choi TG, Nguyen MN, Matondo A,
Jo YH, Yoo JY, Nguyen NN, Yun HR, Kim J, Akter S, et al: An 8-gene
signature for prediction of prognosis and chemoresponse in
non-small cell lung cancer. Oncotarget. 7:86561–86572.
2016.PubMed/NCBI
|
|
7
|
Francois-Vaughan H, Adebayo AO, Brilliant
KE, Parry NMA, Gruppuso PA and Sanders JA: Persistent effect of
mTOR inhibition on preneoplastic foci progression and gene
expression in a rat model of hepatocellular carcinoma.
Carcinogenesis. 37:408–419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Baek SJ, Sato K, Nishida N, Koseki J,
Azuma R, Kawamoto K, Konno M, Hayashi K, Satoh T, Doki Y, et al:
MicroRNA miR-374, a potential radiosensitizer for carbon ion beam
radiotherapy. Oncol Rep. 36:2946–2950. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Paula LM, De Moraes LH, Do Canto AL, Dos
Santos L, Martin AA, Rogatto SR and De Azevedo Canevari R: Analysis
of molecular markers as predictive factors of lymph node
involvement in breast carcinoma. Oncol Lett. 13:488–496.
2017.PubMed/NCBI
|
|
10
|
Li G, Li X, Yang M, Xu L, Deng S and Ran
L: Prediction of biomarkers of oral squamous cell carcinoma using
microarray technology. Sci Rep. 7:421052017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Viziteu E, Klein B, Basbous J, Lin YL,
Hirtz C, Gourzones C, Tiers L, Bruyer A, Vincent L, Grandmougin C,
et al: RECQ1 helicase is involved in replication stress survival
and drug resistance in multiple myeloma. Leukemia. Mar
10–2017.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu J, Wang J and Shen W: Identification of
MAGEA12 as a prognostic outlier gene in gastric cancers. Neoplasma.
64:238–243. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jo J, Nam CM, Sull JW, Yun JE, Kim SY, Lee
SJ, Kim YN, Park EJ, Kimm H and Jee SH: Prediction of colorectal
cancer risk using a genetic risk score: The Korean cancer
prevention study-II (KCPS-II). Genomics Inform. 10:175–183. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ito H, Mo Q, Qin LX, Viale A, Maithel SK,
Maker AV, Shia J, Kingham P, Allen P, DeMatteo RP, et al: Gene
expression profiles accurately predict outcome following liver
resection in patients with metastatic colorectal cancer. PLoS One.
8:e816802013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mármol I, Sánchez-de-Diego C, Dieste
Pradilla A, Cerrada E and Yoldi Rodriguez MJ: Colorectal carcinoma:
A general overview and future perspectives in colorectal cancer.
Int J Mol Sci. 18:pii:E1972017. View Article : Google Scholar
|
|
16
|
Zhu Q, Izumchenko E, Aliper AM, Makarev E,
Paz K, Buzdin AA, Zhavoronkov AA and Sidransky D: Pathway
activation strength is a novel independent prognostic biomarker for
cetuximab sensitivity in colorectal cancer patients. Hum Genome
Var. 2:150092015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu T, Gao YF, Chen YX, Wang ZB, Yin JY,
Mao XY, Li X, Zhang W, Zhou HH and Liu ZQ: Genome-scale analysis
identifies GJB2 and ERO1LB as prognosis markers in patients with
pancreatic cancer. Oncotarget. 8:21281–21289. 2017.PubMed/NCBI
|
|
18
|
Tabas-Madrid D, Nogales-Cadenas R and
Pascual-Montano A: GeneCodis3: A non-redundant and modular
enrichment analysis tool for functional genomics. Nucleic Acids
Res. 40:W478–W483. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gruber-Rouh T, Marko C, Thalhammer A,
Nour-Eldin NE, Langenbach M, Beeres M, Naguib NN, Zangos S and Vogl
TJ: Current strategies in interventional oncology of colorectal
liver metastases. Br J Radiol. May 26–2016.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song N, Shin A, Park JW, Kim J and Oh JH:
Common risk variants for colorectal cancer: An evaluation of
associations with age at cancer onset. Sci Rep. 7:406442017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Slattery ML, Herrick JS, Mullany LE, Gertz
J and Wolff RK: Improved survival among colon cancer patients with
increased differentially expressed pathways. BMC Med. 13:752015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dong G, Mao Q, Yu D, Zhang Y, Qiu M, Dong
G, Chen Q, Xia W, Wang J, Xu L and Jiang F: Integrative analysis of
copy number and transcriptional expression profiles in esophageal
cancer to identify a novel driver gene for therapy. Sci Rep.
7:420602017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao B, Shao Q, Choudhry H, Marcus V, Dong
K, Ragoussis J and Gao ZH: Weighted gene co-expression network
analysis of colorectal cancer liver metastasis genome sequencing
data and screening of anti-metastasis drugs. Int J Oncol.
49:1108–1118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xiong W, Gao D, Li Y, Liu X, Dai P, Qin J,
Wang G, Li K, Bai H and Li W: Genome-wide profiling of
chemoradiation-induced changes in alternative splicing in colon
cancer cells. Oncol Rep. 36:2142–2150. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang H, Jiang X, Wang J, Li Y, Song CX,
Chen P, Li S, Gurbuxani S, Arnovitz S, Wang Y, et al:
Identification of MLL-fusion/MYC-miR-26-TET1 signaling circuit in
MLL-rearranged leukemia. Cancer Lett. 372:157–165. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu XL, Ai ZH, Wang J, Xu YL and Teng YC:
Weighted gene co-expression network analysis in identification of
endometrial cancer prognosis markers. Asian Pac J Cancer Prev.
13:4607–4611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bradley CA, Dunne PD, Bingham V, McQuaid
S, Khawaja H, Craig S, James J, Moore WL, McArt DG, Lawler M, et
al: Transcriptional upregulation of c-MET is associated with
invasion and tumor budding in colorectal cancer. Oncotarget.
7:78932–78945. 2016.PubMed/NCBI
|
|
28
|
Gayyed MF, Abd El-Maqsoud NM, El-Heeny
El-Hameed AA and Mohammed MF: c-MET expression in colorectal
adenomas and primary carcinomas with its corresponding metastases.
J Gastrointest Oncol. 6:618–627. 2015.PubMed/NCBI
|
|
29
|
Raghav K, Morris V, Tang C, Morelli P,
Amin HM, Chen K, Manyam GC, Broom B, Overman MJ, Shaw K, et al: MET
amplification in metastatic colorectal cancer: An acquired response
to EGFR inhibition, not a de novo phenomenon. Oncotarget.
7:54627–54631. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Al-Maghrabi J, Emam E, Gomaa W, Saggaf M,
Buhmeida A, Al-Qahtani M and Al-Ahwal M: c-MET immunostaining in
colorectal carcinoma is associated with local disease recurrence.
BMC Cancer. 15:6762015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Takahashi N, Iwasa S, Taniguchi H, Sasaki
Y, Shoji H, Honma Y, Takashima A, Okita N, Kato K, Hamaguchi T, et
al: Prognostic role of ERBB2, MET and VEGFA expression in
metastatic colorectal cancer patients treated with anti-EGFR
antibodies. Br J Cancer. 114:1003–1011. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jia Y, Dai G, Wang J, Gao X, Zhao Z, Duan
Z, Gu B, Yang W, Wu J, Ju Y, et al: c-MET inhibition enhances the
response of the colorectal cancer cells to irradiation in
vitro and in vivo. Oncol Lett. 11:2879–2885.
2016.PubMed/NCBI
|
|
33
|
Sun Y, Sun L, An Y and Shen X:
Cabozantinib, a Novel c-Met Inhibitor, inhibits colorectal cancer
development in a Xenograft model. Med Sci Monit. 21:2316–2321.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lu D, Yao Q, Zhan C, Le-Meng Z, Liu H, Cai
Y, Tu C, Li X, Zou Y and Zhang S: MicroRNA-146a promote cell
migration and invasion in human colorectal cancer via
carboxypeptidase M/src-FAK pathway. Oncotarget. 8:22674–22684.
2017.PubMed/NCBI
|
|
35
|
Denis CJ and Lambeir AM: The potential of
carboxypeptidase M as a therapeutic target in cancer. Expert Opin
Ther Targets. 17:265–279. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang B, Wang W, Zhu Z, Zhang X, Tang F,
Wang D, Liu X, Yan X and Zhuang H: Mitochondrial serine
hydroxymethyltransferase 2 is a potential diagnostic and prognostic
biomarker for human glioma. Clin Neurol Neurosurg. 154:28–33. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang L, Chen Z, Xue D, Zhang Q, Liu X,
Luh F, Hong L, Zhang H, Pan F, Liu Y, et al: Prognostic and
therapeutic value of mitochondrial serine
hydroxyl-methyltransferase 2 as a breast cancer biomarker. Oncol
Rep. 36:2489–2500. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim D, Fiske BP, Birsoy K, Freinkman E,
Kami K, Possemato RL, Chudnovsky Y, Pacold ME, Chen WW, Cantor JR,
et al: SHMT2 drives glioma cell survival in ischaemia but imposes a
dependence on glycine clearance. Nature. 520:363–367. 2015.
View Article : Google Scholar : PubMed/NCBI
|