Introduction
Circadian rhythms control the 24 h cycle of specific
metabolic functions required by living beings, ensuring an
efficient metabolic homeostasis (1,2). In
humans, the circadian rhythms are controlled by a master pacemaker
situated in the suprachiasmatic nuclei of the hypothalamus, which
is synchronized to the photoperiod (1,2). The
molecular clock of transcription involves a translational feedback
mechanism of genes, including clock circadian regulator (CLOCK),
period circadian clock (PER), aryl hydrocarbon receptor nuclear
translocator like (BMAL1) and cryptochrome circadian clock (CRY),
able to regulate a number of physiological properties, including
body temperature, melatonin secretion, hormone secretion, blood
pressure and the sleep-wake cycle (3).
It is well-known that disturbances in the circadian
rhythm may cause the development of diseases, including major
depressive disorder, seasonal affective disorder, schizophrenia,
bipolar disorder (4–9), stress, desynchronosis (9), anxiety disorder, diabetes (10), obesity, diseases associated with aging
(11), genome instability (12) and cancer (13,14). Prior
studies have demonstrated the association between circadian rhythm
alterations and the development of breast (15) and prostate cancer (16), B-cell lymphoma (17), non-small cell lung (18), testicular (19) and ovarian cancer (20).
The liver has a central and unique metabolic
function in maintaining energy homeostasis via glycolysis and
gluconeogenesis associated with fatty acid metabolism
(biosynthesis/beta oxidation) (21).
Rhythmic fluctuations have been identified in hepatic metabolic
functions with a 24-h periodicity (22). Previous studies have demonstrated that
liver cancer initiation may be due to alterations in circadian
rhythmic genes, including PER3 (23)
and CRY genes, and casein kinases (24). Additionally, it has been revealed that
the dysregulation of metallothionein-1 (MT-1), MT-2 and metal
transcription factor-1 are involved in the alterations to circadian
rhythms present in liver cancer (25). Furthermore, the presence of hepatitis
viral infection, which is a cause of liver cancer, has been
revealed to cause dysregulation of the expression of circadian
genes (26). However, it is crucial
to underline that liver cancer is additionally caused by chronic
exposure to toxic chemicals, hepatosteatosis, type 2 diabetes and
obesity (27–29). The underlying molecular mechanism of
circadian clock disruption in non-viral liver cancer remains
unknown.
Our previous study analyzed the transcriptome of a
human line of hepatoblastoma cells without viral infection (HepG2),
compared with normal hepatocytes, and the gene expression data,
obtained from the liver tissues of patients with hepatitis C virus
(HCV), HCV-associated cirrhosis and liver cancer with
HCV-associated cirrhosis, using the publicly available E-MTAB-950
database (30). Despite the HepG2
cell line being revealed as a misidentified hepatocellular
carcinoma cell line and subsequently identified as hepatoblastoma
by the International Cell Line Authentication Committee (iclac.org/databases/cross-contaminations), the
outcomes of our studies are validated because they focused on a
liver cancer cell line without viral infection. Our previous
analysis enabled the identification of specific clusters of genes
for the various stages of liver cancer, and allowed the isolation
of a network of 26 HUB genes that specifically control critical
metabolic functions, independent of the viral infection (30). All 26 HUB genes were revealed to
encode intrinsically disordered proteins (IDPs), thus they
exhibited multifunctional behaviors and were involved in metabolic
cellular control.
The present study aimed to identify whether these
HUB genes (and associated proteins) were components of the network
that controls the human circadian rhythm. Therefore, the network of
genes involved in circadian rhythms was extracted from the human
interactome to identify the nodes with high centrality and
interactions with the most studied circadian gene, CLOCK.
Furthermore, the association between the circadian network and 26
HUB genes in the HepG2 network was evaluated. In this way, we
identified the genes linking the two main networks and evaluated if
the proteins coded from them have high disorder propensities.
Finally, comparing the deregulated microRNA (miRNA/miR) in HepG2
cells with miRNAs in normal hepatocytes, the target genes were
predicted in order to evaluate if there were miRNAs involved in the
network between HepG2 cells and circadian networks.
Materials and methods
Network analysis
A network of 71 genes was extracted, from the human
interactome compiled from various databases, including Pathway
Commons (31), Biological General
Repository for interaction Datasets (BioGRID) (32), Human Protein Reference Database
(33), ConsensusPathDB (34), Database of Interacting Proteins
(35) and the Breast Cancer
Information Core and Michigan Molecular Interactions (36), and identified to be involved in
circadian rhythms. Only the connected component of these 71 seed
networks were considered for statistical analysis using different
tools, including Network-Analyzer (37), the Database for Annotation,
Visualization and Integrated Discovery (DAVID) (38) and the Biological Networks Gene
Ontology tool (39). Using the
Cytoscape 3.5 package (www.cytoscape.org), statistical analysis was performed
to evaluate the following three measures of centrality: i) The
degree, which indicates the number of interactions of a particular
node with other nodes in the network; ii) the betweenness, which
evaluates the importance of a node in the network and how the other
interactions in the network are controlled by this node (40); iii) the closeness centrality of a
node, which measures the speed of information flow through this
node to reachable nodes in the network and ranges from 0–1
(41). However, the power law is used
to predict the HUB nodes that have functions in the network. The
power law details the functional association between two
quantities, where one quantity varies as a power of another; based
on the power law distribution degree, a network may be defined as
scale-free indicating that ‘riches get richer’ (42–44).
Other topological analysis, including average
characteristic path length, network density, centralization and
heterogeneity were evaluated using the Cytoscape 3.5. package
(41,45). The characteristic path length is
calculated by identifying the shortest path between all pairs of
nodes, adding them and dividing by the total number of pairs. This
indicates the number of steps required to get from one member of
the network to another (41,45). The density of a network is defined as
a ratio of the number of edges to the number of possible edges
(46); whereas, the centralization
produces rankings identify the most important nodes in a network
model (47). In particular, networks
with topologies resembling a star have a centralization close to 1,
whereas decentralized networks are characterized by having a
centralization of ~0.47. Furthermore, the network heterogeneity,
evaluated using the Cytoscape 3.5. package, reflects the tendency
of a network to contain HUB nodes (48). Finally, a cluster analysis was
performed, which groups similar objects to form clusters;
therefore, objects in the same cluster are more similar to each
other, compared with those in other clusters. In particular, the
overlapping clusters were calculated on the basis of cohesiveness
quality functions (49).
Disorder propensity analysis
The associated protein sequences corresponding to
the HUB genes common between circadian and liver cancer networks
were extracted from the UniProt database (www.uniprot.org). To assess the proportion of residues
involved in intrinsic disorder, the DisProt tool (50) was used to subdivide the sequences into
three major groups extracted on the basis of similar contents of
disorder (10–15%, 16–50% and >50%).
Target genes prediction of miRNAs
A list of miRNAs (51)
that have been identified as dysregulated in HepG2 cells, compared
with human normal hepatocytes were selected. Furthermore,
predictions of miRNA complementarity to 3′ untranslated regions
(UTRs) in mRNAs were performed using three commonly used tools for
target prediction: TargetScan Human 6.2 (www.targetscan.org) (52), PITA (53) and miRanda (www.microrna.org) (54).
This analysis was based on identifying conserved sites that match
the seed region of each miRNA, corresponding to the position
between nucleotides 2 and 8in mature miRNAs. A list of putative
targets for each miRNA was obtained and those predicted from 2/3
tools were selected for functional annotation analysis of pathways,
which was performed using the DAVID program and by selecting the
more significantly enriched pathways, with a number of genes >60
and P<0.05 (38).
Results and discussion
Human circadian network. The network of genes
involved in circadian rhythms, on the basis of seed nodes, was
extracted from the human interactome, and included the following
genes: Aryl Hydrocarbon Receptor Nuclear Translocator Like (ARNTL),
casein kinase (CSNK)1E, inter-α-trypsin inhibitor heavy chain
family member 5 (ITIH5), replication factor C subunit 3 (RFC3), WD
repeat domain 41, PER1, CSNK1D, syntrophinβ2 (SNTB2), acyl-CoA
thioesterase 13, chondroitin sulfate
N-acetylgalactosaminyltransferase 1, PER2, ARNTL2, PDZ domain
containing ring finger 3, growth arrest specific 2, fibronectin
leucine rich transmembrane protein 1, PER3, neuronal PAS domain
protein 2, low density lipoprotein receptor, zinc finger protein
(ZNF) 286A, G protein-coupled receptor (GPR)116, nuclear receptor
subfamily 1 group D member 1, cyclin dependent kinase (CDK) L5,
splicing factor proline and glutamine rich (SFPQ), adipogenesis
regulatory factor, translocase of inner mitochondrial membrane 8A,
basic helix-loop-helix family member E (BHLHE)40,
7-dehydrocholesterol reductase, solute carrier family 39 member 14,
suppressor of cytokine signaling 2 (SOCS2), GPR6, BHLHE41, histone
deacetylase 4 (HDAC4), HLF PAR bZIP transcription factor (HLF),
solute carrier organic anion transporter family member 4A1,
γ-secretase activating protein, BMAL1, methyl-CpG binding protein 2
(MECP2), ETS variant 5 (ETV5), Kruppel like factor 11 (KLF11),
ZNF394, D-box binding PAR bZIP transcription factor, neurexin 1,
TNFAIP3 interacting protein 2 (TNIP2), exocyst complex component 1
(EXOC1), extended synaptotagmin 1 (ESYT1), nuclear receptor
subfamily 1 group D member 2, SH3 and multiple Ankyrin repeat
domains 3 (SHANK3), zw10 kinetochore protein (ZW10), phospholipid
scramblase 1 (PLSCR1), CLOCK, solute carrier family 2 member 1
(SLC2A1), hydrocretin receptor 2,
5-methyltetrahydrofolate-homocysteine methyltransferase (MTR),
transferrin receptor (TFRC), casein kinase 1-α-1, synemin, sprouty
RTK signaling antagonist 4 (SPRY4), ubiquitin specific peptidase 2,
glycogen synthase kinase 3β (GSK3B), hypoxia inducible lipid
droplet associated, Scmpolycomb group protein like 1, CRY1, nuclear
factor interleukin 3 regulated, ATPase H+/K+ transporting α
subunit, Ras homolog family member B, CRY2, insulin induced gene 1,
unc-13 homolog A (UNC13A) and apolipoprotein L domain containing 1
(APOLD1). The human circadian network consists of 2151 nodes and
75821 interactions (Table I). The
circadian network was identified to be highly centralized (0.235);
a higher value of centralization indicates that the network is
concentrated in the center with an overall integration towards the
high degree nodes. The network density of the circadian network,
which describes the proportion of potential connections in a
network that are actual connections, as a measure of network
effectiveness, was identified to be 0.033. In addition, the
circadian network exhibited a high value of heterogeneity, which
demonstrates its tendency to contain HUB nodes. The characteristic
path length was identified to be 2.373, whereas the average number
of neighbors was 70.5.
 | Table I.Statistical analysis of the genes
involved in the network obtained for human circadian rhythms. |
Table I.
Statistical analysis of the genes
involved in the network obtained for human circadian rhythms.
| Statistical
analysis | Circadian
network |
|---|
| Number of
nodes | 2151 |
| Number of
interactions | 75821 |
| Network
density | 0.033 |
| Network
centralization | 0.235 |
| Characteristic path
length | 2.373 |
| Network
heterogeneity | 1.012 |
| Neighbor average
number | 70.498 |
The human circadian network was demonstrated to
follow the small-world rule (41), as
the characteristic path length is very short. The nodes that
exhibited a high centrality were small ubiquitin-like modifier 2,
CDK2, heat shock protein 90A, p53, nuclear respiratory factor 1 and
GSK3B. As CLOCK is one of the most studied circadian genes, its
sub-network was extracted from the general network of circadian
genes using the Cytoscape tool, which demonstrated that it
contained 87 nodes with 86 direct interactions. The analysis of the
present study demonstrated that CLOCK is associated with other
genes including proliferating cell nuclear antigen, PER and sirtuin
1, 3 and 5.
Association between the circadian network
and 26 HUBs in the HepG2 network
Four networks have previously been compared, each
obtained from the differentially expressed genes in HepG2 cells and
in liver tissues from patients with HCV, HCV-associated cirrhosis
and liver cancer with HCV-associated cirrhosis, using the publicly
available E-MTAB-950 with the entire human interactome as the
background (30,55). The aim was to discriminate between
liver cancer in the presence or absence of viral infection, and to
identify the presence and the function of common or specific HUB
nodes in the four networks. Although HepG2 cells were revealed to
be misidentified as a hepatocellular carcinoma cell line instead a
hepatoblastoma by International Cell Line Authentication Committee
(iclac.org/databases/cross-contaminations), the results
obtained using this cell line were used, as the study focused on a
liver cancer cell line without viral infection. In the present
study, it was evaluated whether these specific genes in HepG2 cells
were components of the circadian rhythm, by identifying their
presence in the circadian network. The results of the present study
demonstrated that 20/26 HUB genes [CSNK2α1, SH2 domain containing
(SRC), ubiquitin D, aurora kinase B (AURKB), cytoskeleton
associated protein 5 (CKAP5), replication factor C subunit 4, cell
division cycle 20 (CDC20), stratifin, minichromosomemaintenance
complex component (MCM)6, checkpoint kinase 1, centromere protein A
(CENPA), HLA-B, baculoviral IAP repeat containing 5 (BIRC5), MCM3,
mitotic arrest deficient 2 like 1 (MAD2L1), MCM4, ZW10 interacting
kinetochore protein (ZWINT), kinesin family member (KIF)2C, inner
centromere protein (INCENP) and SPC24 NDC80 kinetochore complex
component (SPC24)] were demonstrated to be components of the human
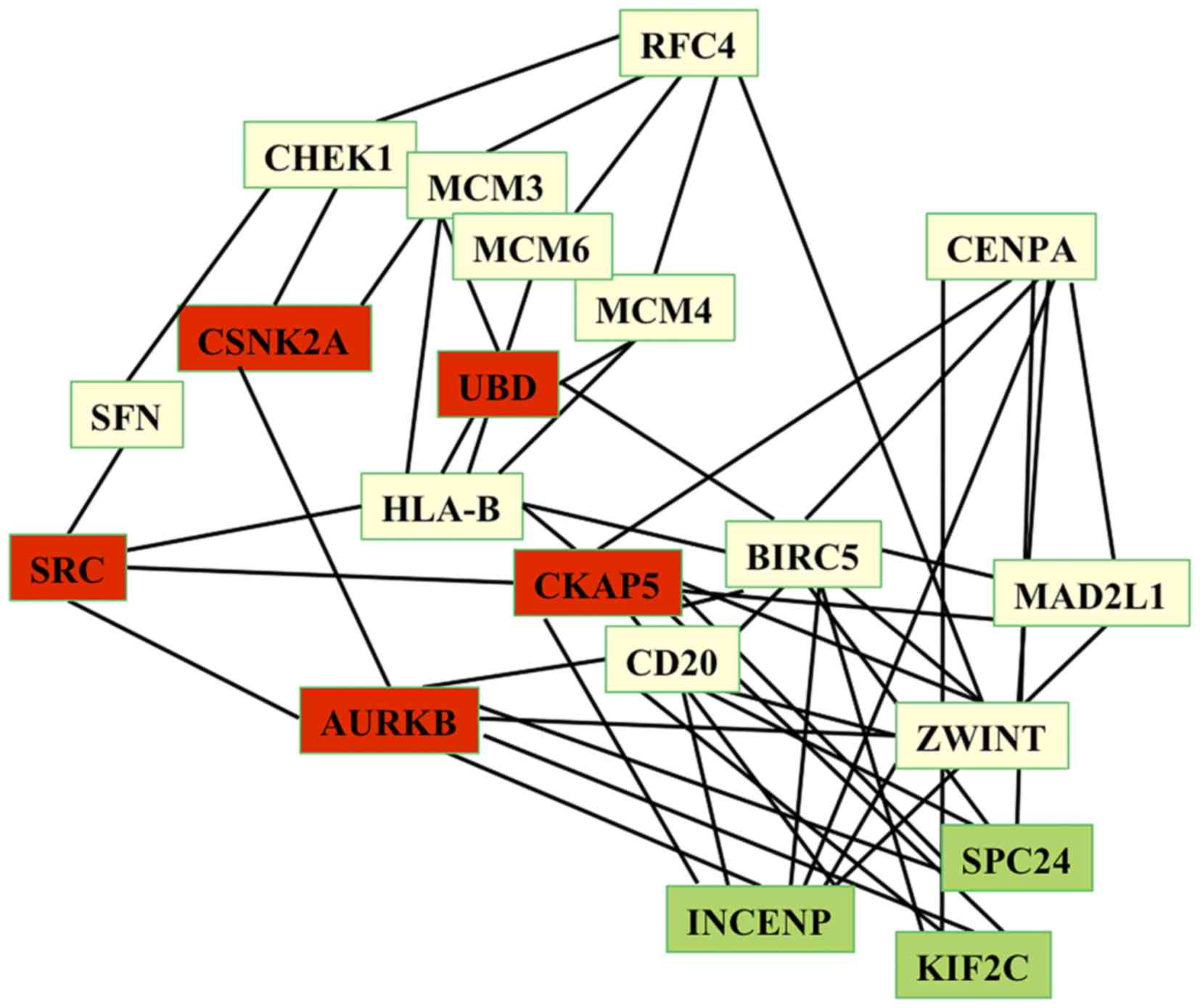
circadian network (Fig. 1).
The aforementioned 20 HUB genes revealed high degree
values in the circadian network, ranging between 287 and 77,
indicating that these genes control a large number of metabolic
functions and the flow of information via the circadian network.
Furthermore, the results of the present study demonstrated that
these 20 HUB genes interacted with 31 seed circadian genes,
including CLOCK, PER1-3, CRY1-2, ARNTL2, CSNK1D, HDAC4, ZW10,
CSNK1E, RFC3, MTR, SFPQ, ESYT1, transferrin receptor, GSK3B, EXOC1,
SHANK3 and PLSCR1. In particular, our study has revealed that CLOCK
is associated with HUB genes of the HepG2 network via CKAP5, which
exhibits a high degree (217), a short path length value of 2.053
and a high value of stress centrality. Thus, CKAP5 interacts via a
number of the shortest path-lengths of the network, which makes it
a perfect link between circadian and HepG2 networks.
CKAP5 encodes a cytoskeleton-associated protein
belonging to the TOG/XMAP215 family, and CKAP is also known as a
colonic and hepatic tumor overexpressed gene protein (56,57). Its
coded protein serves two distinct functions in spindle formation
and in the protection of kinetochore microtubules, via the control
of the de-polymerization process and centrosomal microtubule
assembly (58,59). These two processes regulate the
mitotic cell cycle via spindle formation (60) and the interaction between microtubules
and the cell cortex for the directional cell movement. Notably, the
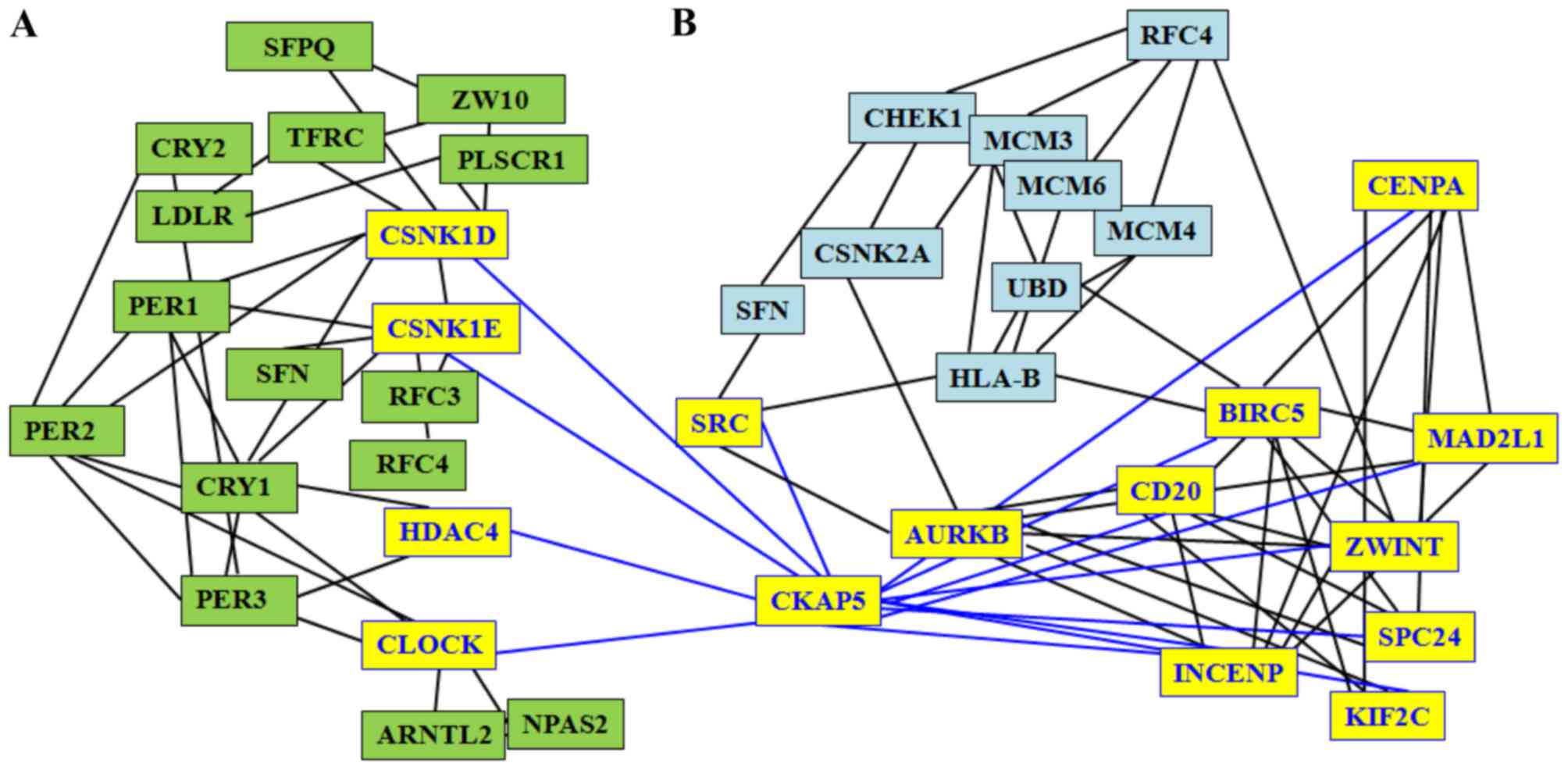
results of the present study identified that CKAP5 is associated
with three circadian genes (CSNK1E, CSNK1D and HDAC4) and with 10
HepG2 genes (SRC, ZWINT, AURKB, CDC20, CENPA, INCENP, MAD2L1,
BIRC5, SPC24 and KIF2C; Fig. 2).
Therefore, the two sub-networks of circadian genes and HepG2 genes
connected via CKAP5 may be disturbed by any alteration associated
with circadian and liver cancer genes. Due to the close
associations between nodes, a putative perturbing stressor, for
example an alteration of a circadian gene, can induce a
perturbation to the global network, and thus, cancer progression
(61).
Structural analysis on common nodes between
HepG2 and circadian networks
As all HUB genes identified in our network analysis
encode for proteins, it is crucial to understand whether the genes
possess specific structural features. Previously, it was
demonstrated that the metabolic sub-network specific for liver
cancer is formed only by IDPs (30,55).
Structural flexibility and binding plasticity enable IDPs to
interact with a broad range of molecular partners (30,55).
Therefore, in the present study, disorder propensity and the number
of molecular partners, with which these proteins can interact, was
evaluated using DisProt and BioGRID tools, respectively. As
presented in Table II, the proteins
encoded by the genes common to the circadian and HepG2 networks
belonged to the IDP family. In particular, 3, 11 and 7 proteins
exhibited 15%, 16–50% and >50% ID regions (IDRs), respectively.
Subsequently, the physical interactions between all proteins
encoded by the genes common to the circadian and HepG2 network and
other proteins with which they can interact were analyzed, and
demonstrated that they are able to form between 17 and 667
interactions. This result supports our hypothesis that the
flexibility of the disordered regions functions in the
establishment a high number of interactions.
 | Table II.Molecular properties of the 20 HUB
nodes specific for HepG2 that are common to the circadian
network. |
Table II.
Molecular properties of the 20 HUB
nodes specific for HepG2 that are common to the circadian
network.
| Gene name | Protein code | Protein name | IDR | INT | SEQ |
|---|
| AURKB | Q96GD4 | Aurora kinase
B | ++ | 268 | 344 |
| BIRC5 | O15392 | Baculoviral IAP
repeat-containingprotein 5 | +++ | 137 | 142 |
| CDC20 | Q12834 | Cell division cycle
protein 20 homolog | ++ | 595 | 499 |
| CENPA | P49450 | Histone H3-like
centromeric protein A | ++ | 129 | 140 |
| CHEK1 | O14757 |
Serine/threonine-protein kinase Chk1 | ++ | 161 | 476 |
| CKAP5 | Q14008 | Cytoskeleton
associated protein 5 | +++ | 39 | 2032 |
| CSNK2A1 | P68400 | Casein kinase II
subunit alpha | ++ | 599 | 391 |
| HLA-B | P01889 | HLA class I
histocompatibility antigen, B-7 alpha chain | ++ | 63 | 362 |
| INCENP | Q9NQS7-INCE
human | Inner centromere
protein | +++ | 17 | 918 |
| KIF2C | Q99661 | Kinesin-like
protein KIF2C | ++ | 82 | 725 |
| MAD2L1 | Q13257 | Mitotic spindle
assembly check point protein MAD2A | + | 48 | 205 |
| MCM3 | P25205 | DNA replication
licensing factor MCM 3 | ++ | 171 | 808 |
| MCM4 | P33991 | DNA replication
licensing factor MCM 4 | ++ | 152 | 863 |
| MCM6 | Q14566 | DNA replication
licensing factor MCM 6 | ++ | 160 | 821 |
| RFC4 | P35249 | Replication factor
C subunit 4 | + | 68 | 363 |
| SFN | P31947 | 14-3-3 protein
sigma | +++ | 305 | 248 |
| SPC24 | Q8NBT2 | Kinetochore protein
Spc24 | +++ | 31 | 197 |
| SRC | P12931 | Proto-oncogene
tyrosine-protein kinase Src | ++ | 272 | 536 |
| UBD | 15205 | Ubiquitin D | +++ | 667 | 165 |
| ZWINT | 95229 | Zw10-
interactor | + | 55 | 277 |
Association between miRNAs and genes of the
human circadian and HepG2 networks
Following the identification of the associations
between circadian and HepG2 networks, and demonstrating that genes
common to the two networks encode IDPs, the presence of miRNAs
involved in the sub-network between HepG2 and circadian genes was
evaluated. A prior study identified the deregulated miRNAs in HepG2
cells compared with normal hepatocytes (51). A total of 11 downregulated
(miR-146b-5p, miR-195, miR-122, miR-122a, miR-375, miR-885-5p,
miR-768-5p, miR-101, miR-192, miR-194 and miR-215) and 2
upregulated miRNAs (miR-221 and miR-99b) were identified (51). Therefore, in the present study, the
target genes of these 13 miRNAs were predicted using the three
aforementioned tools. The results of the present study identified
5415 target genes belonging to different metabolic pathways
including axon guidance, hippo signaling, endocytosis,
phosphoinositide 3-kinase/protein kinase B (AKT) signaling, RAS
signaling, RAP1 signaling and chemokine signaling.
To determine the target genes involved in the HepG2
and circadian networks, genes common to the identified 5,415 target
genes and the circadian genes were identified. The results of the
present study identified 28 genes (APOLD1, BHLHE40, BHLHE41, CLOCK,
CRY1, CRY2, extended synaptotagmin 1, ETV5, HDAC4, HLF, ITIH5,
KLF11, MECP2, MTR, nuclear receptor subfamily 2 group D member 2,
PER1, PER2, PER3, RFC3, SLC2A1, SNTB2, SOCS2, SPRY4, TFRC, TNIP2,
UNC13a and ZW10) that are common between circadian genes and the
5415 identified target genes. In addition, four genes (checkpoint
kinase 1, KIF2C, MCM6 and MAD2L1), that associated the HepG2
network with the circadian network, were targeted by the following
four miRNAs: miR-195-5p, miR-192-5p, miR-122-5p and miR-101-3p. The
present study identified three genes (SNTB2, HLF and CRY2) that
correlated four miRNAs (miR-195-5p, miR-192-5p, miR-122-5p and
miR-101-3p) between them (Fig.
3).
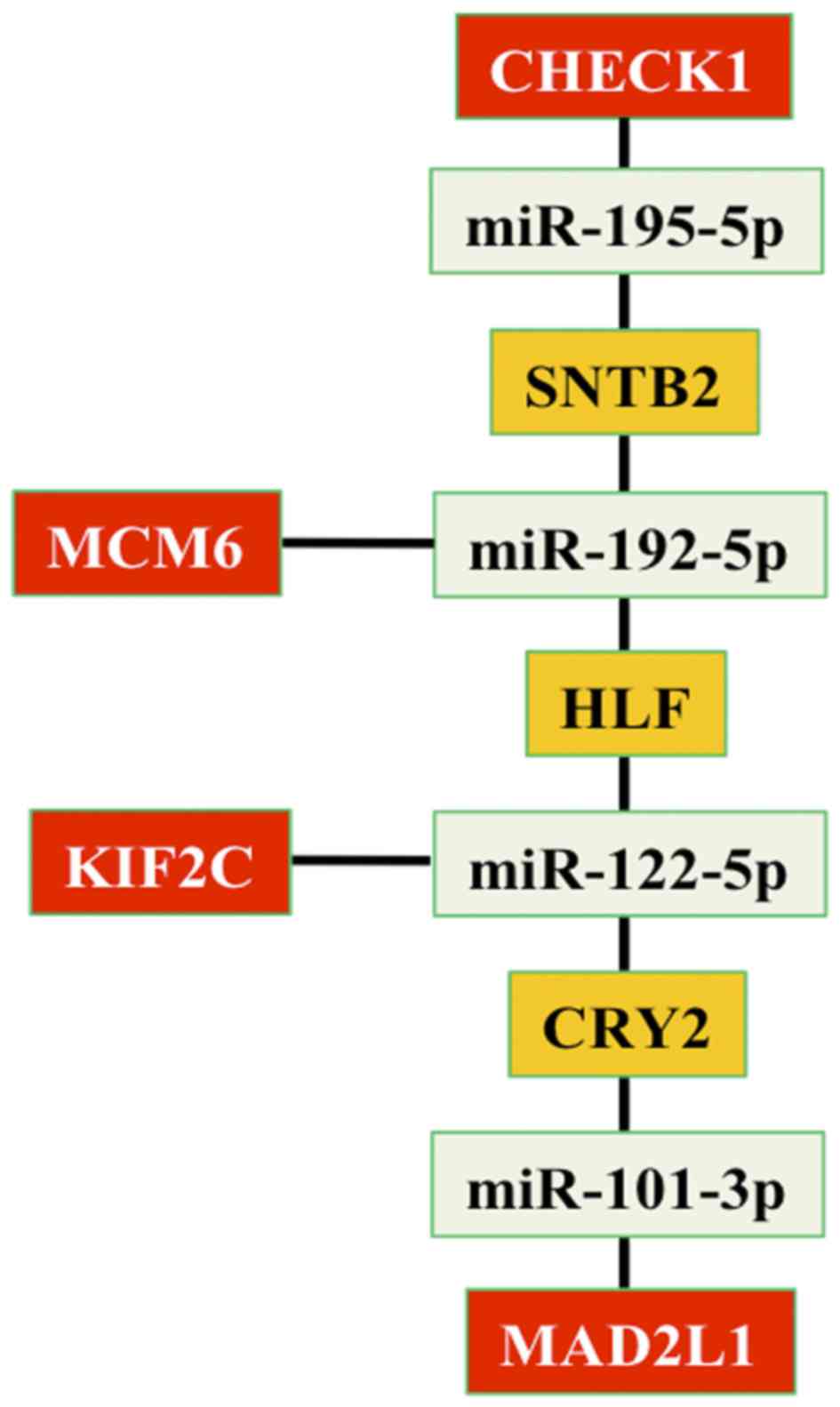 | Figure 3.Associations between genes and miRNAs
that link HepG2 and circadian networks. In details, four genes
(CHECK1, KIF2C, MCM6 and MAD2L1) are reported in red; four miRNAs
(miR-195-5p, miR-192-5p, miR-122-5p and miR-101-3p) are reported in
green and three genes (SNTB2, HLF and CRY2) that correlate the
miRNAs are reported in orange. miR, microRNA; SNTB2, syntrophin β2;
MCM6, minichromosome maintenance complex component 6; HLF, HLF PAR
bZIP transcription factor; KIF2C, kinesin family member 2C; CRY2,
cryptochrome circadian clock 2; MAD2L1, mitotic arrest deficient 2
like 1; CHECK1, checkpoint kinase 1. |
Previous studies have suggested that liver cancer is
associated with abnormalities in circadian rhythms (26,62) due to
alterations in the expression of certain circadian genes in
cancerous cells, induced by hypoxia (24), or the overexpression of the mammalian
timeless protein (23), a protein
that controls chromosome integrity, growth and development. The
present study was, to the best of our knowledge, the first to
identify a sub-set of HUB genes consisting of genes present in the
HepG2 cell network, and involved in cancer progression, in common
with human circadian rhythm genes. The results of the present study
revealed the following: i) CLOCK is associated, via CKAP5, with the
HUB genes of the HepG2 network; ii) CKAP5 is associated with three
other circadian genes (CSNK1E, CSNK1D and HDAC4), and with 10HepG2
genes (SRC, ZWINT, AURKB, CDC20, CENPA, INCENP, MAD2L1, BIRC5,
SPC24 and KIF2C); iii) the genes linking the circadian system and
liver cancer codify for proteins that exhibit IDRs; iv) a sub-panel
of seven genes and three miRNAs link human circadian rhythms with
liver cancer in a single network.
Sassoni-Corsi et al (63) demonstrated that the liver operates as
an exclusive endogenous metabolic reorganizer in tumor-bearing mice
(63). Notably, associations between
cancer and circadian genes are maintained while the
pro-inflammatory response of the liver is altered and leads to the
disturbance of AKT, AMP-activated protein kinase and sterol
regulatory element binding protein signaling, which, in turn,
affects glucose and lipid metabolism. These results demonstrate the
requirement to study associations between the circadian rhythms in
liver cancer and/or other types of cancer (63). In this context, the results of the
present study indicated that further studies are required to
determine how the structural perturbation of the HUB nodes in liver
cancer may trigger significant and widespread sources of functional
changes, which consequently may produce the distinct metabolic
functions of cancer cells.
References
|
1
|
Czeisler CA and Klerman EB: Circadian and
sleep-dependent regulation of hormone release in humans. Recent
ProgHorm Res. 54:97–130. 1999.
|
|
2
|
Dibner C and Schibler U: Circadian timing
of metabolism in animal models and humans. J Intern Med.
277:513–527. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Harfmann BD, Schroder EA and Esser KA:
Circadian rhythms the molecular clock and skeletal muscle. J Biol
Rhythms. 30:84–94. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
West AC and Bechtold DA: The cost of
circadian desynchrony: Evidence insights and open questions.
BioEssays. 37:777–788. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fares S, Hermens DF, Naismith SL, White D,
Hickie IB and Robillard R: Clinical correlates of chronotypes in
young persons with mental disorders. Chronobiol Int. 32:1183–1191.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Geoffroy PA, Etain B, Sportiche S and
Bellivier F: Circadian biomarkers in patients with bipolar
disorder: Promising putative predictors of lithium response. Int J
Bipolar Disord. 2:282014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Geoffroy PA, Etain B, Franchi JA,
Bellivier F and Ritter P: Melatonin and melatonin agonists as
adjunctive treatments in bipolar disorders. Curr Pharm Des.
21:3352–3358. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Benedetti F, Riccaboni R, Dallaspezia S,
Locatelli C, Smeraldi E and Colombo C: Effects of CLOCK gene
variants and early stress on hopelessness and suicide in bipolar
depression. Chronobiol Int. 32:1156–1161. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Russo M, Mahon K, Shanahan M, Ramjas E,
Solon C, Purcell SM and Burdick KE: The relationship between sleep
quality and neurocognition in bipolar disorder. J Affect Disord.
187:156–162. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Challet E: Keeping circadian time with
hormones. Diabetes Obes Metab. 17:76–83. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Anisimov VN, Vinogradova IA, Panchenko AV,
Popovich IG and Zabezhinski MA: Light-at-night-induced circadian
disruption cancer and aging. Curr Aging Sci. 5:170–177. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Belancio VP: LINE-1 activity as molecular
basis for genomic instability associated with light exposure at
night. Mob Genet Elements. 5:1–5. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Savvidis C and Koutsilieris M: Circadian
rhythm disruption in cancer biology. Mol Med. 18:1249–1260. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Uth K and Sleigh R: Deregulation of the
circadian clock constitutes a significant factor in tumorigenesis:
A clockwork cancer. Part II In vivo studies. Biotechnol Biotechnol
Equip. 28:379–386. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Michael AK, Harvey SL, Sammons PJ,
Anderson AP, Kopalle HM, Banham AH and Partch CL: Cancer/Testis
antigen PASD1 silences the circadian clock. Mol Cell. 58:743–754.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kiss Z and Ghosh PM: Woman in cancer
thematic review: Circadian rhythmicity and the influence of ‘clock’
genes on prostate cancer. Endocr Relat Cancer. 23:T123–T134. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gutiérrez-Monreal MA, Villela L, Baltazar
S, Perfecto-Avalos Y, Cardineau GA and Scott SP: A PER3
polymorphism is associated with better overall survival in diffuse
large B-cell lymphoma in Mexican population. Cancer Biomark.
15:699–705. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li J, Chen R, Ji M, Zou SL and Zhu LN:
Cisplatin-based chronotherapy for advanced non-small cell lung
cancer patients: A randomized controlled study and its
pharmacokinetworkics analysis. Cancer Chemother Pharmacol.
76:651–655. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mitchell MI and Engelbrecht AM: Circadian
rhythms and breast cancer: The role of Per2 in doxorubicin-induced
cell death. J Toxicol. 2015:3923602015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Poole EM, Schernhammer E, Mills L,
Hankinson SE and Tworoger SS: Urinary melatonin and risk of ovarian
cancer. Cancer Causes Control. 26:1501–1506. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schmutz I, Albrecht U and Ripperger JA:
The role of clock genes and rhythmicity in the liver. Mol Cell
Endocrinol. 349:38–44. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vollmers C, Gill S, DiTacchio L,
Pulivarthy SR, Le HD and Panda S: Time of feeding and the intrinsic
circadian clock drive rhythms in hepatic gene expression. ProcNatl
Acad Sci USA. 106:21453–21458. 2009. View Article : Google Scholar
|
|
23
|
Elgohary N, Pellegrino R, Neumann O,
Elzawahry HM, Saber MM, Zeeneldin AA, Geffers R, Ehemann V,
Schemmer P, Schirmacher P and Longerich T: Protumorigenic role of
timeless in hepatocellular carcinoma. Int J Oncol. 46:597–606.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu C, Yang SL, Fang X, Jiang JX, Sun CY
and Huang T: Hypoxia disrupts the expression levels of circadian
rhythm genes in hepatocellular carcinoma. Mol Med Rep.
11:4002–4008. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li H, Lu YF, Chen H and Liu J:
Dysregulation of metallothionein and circadian genes in human
hepatocellular carcinoma. Chronobiol Int. 20:192–202. 2016.
|
|
26
|
Yang SL, Yu C, Jiang JX, Liu LP, Fang X
and Wu C: Hepatitis B virus X protein disrupts the balance of the
expression of circadian rhythm genes in hepatocellular carcinoma.
Oncol Lett. 8:2715–2720. 2014.PubMed/NCBI
|
|
27
|
Turati F, Talamini R, Pelucchi C, Polesel
J, Franceschi S, Crispo A, Izzo F, La Vecchia C, Boffetta P and
Montella M: Metabolic syndrome and hepatocellular carcinoma risk.
Br J Cancer. 108:222–658. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Polesel J, Zucchetto A, Montella M, Dal
Maso L, Crispo A, La Vecchia C, Serraino D, Franceschi S and
Talamini R: The impact of obesity and diabetes mellitus on the risk
of hepatocellular carcinoma. Ann Oncol. 20:353–357. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shivappa N, Hébert JR, Polesel J,
Zucchetto A, Crispo A, Montella M, Franceschi S, Rossi M, La
Vecchia C and Serraino D: Inflammatory potential of diet and risk
for hepatocellular cancer in a case-control study from Italy. Br J
Nutr. 115:324–331. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Singh S, Colonna G, Di Bernardo G,
Bergantino F, Cammarota M, Castello G and Costantini S: The gene
expression profiling of hepatocellular carcinoma by a network
analysis approach shows a dominance of intrinsically disordered
proteins (IDPs) between hub nodes. Mol Biosyst. 11:2933–2945. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cerami EG, Gross BE, Demir E, Rodchenkov
I, Babur O, Anwar N, Schultz N, Bader GD and Sander C: Pathway
commons a web resource for biological pathway data. Nucleic Acids
Res. 39:(Database issue). D685–D690. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stark C, Breitkreutz BJ, Reguly T, Boucher
L, Breitkreutz A and Tyers M: BioGRID: A general repository for
interaction datasets. Nucleic Acids Res. 34:(Database issue).
D535–D539. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Peri S, Navarro JD, Amanchy R, Kristiansen
TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B,
Gandhi TK, Gronborg M, et al: Development of human protein
reference database as an initial platform for approaching systems
biology in humans. Genome Res. 13:2363–2371. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kamburov A, Wierling C, Lehrach H and
Herwig R: ConsensusPathDB-a database for integrating human
functional interaction networks. Nucleic Acids Res. 37:(Database
issue). D623–D628. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xenarios I, Salwinski L, Duan XJ, Higney
P, Kim SM and Eisenberg D: DIP, the database of interacting
proteins: A research tool for studying cellular networks of protein
interactions. Nucleic Acids Res. 30:303–305. 2012. View Article : Google Scholar
|
|
36
|
Jayapandian M, Chapman A, Tarcea VG, Yu C,
Elkiss A, Ianni A, Liu B, Nandi A, Santos C, Andrews P, et al:
Michigan Molecular Interactions (MiMI): Putting the jigsaw puzzle
together. Nucleic Acids Res. 35:(Database issue). D566–D571. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cline MS, Smoot M, Cerami E, Kuchinsky A,
Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross
B, et al: Integration of biological networks and gene expression
data using Cytoscape. Nat Protoc. 2:2366–2382. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Maere S, Heymans K and Kuiper M: BiNGO: A
cytoscape plugin to assess overrepresentation of gene ontology
categories in biological networks. Bioinformatics. 21:3448–3469.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yoon J, Blumer A and Lee K: An algorithm
for modularity analysis of directed and weighted biological
networks based on edge-betweenness centrality. Bioinformatics.
22:3106–3108. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Newman MEJ: A measure of betweenness
centrality based on random walks. Soc Networks. 27:39–54. 2005.
View Article : Google Scholar
|
|
42
|
Barabási AL, Gulbahce N and Loscalzo J:
Network medicine: A network-based approach to human disease. Nat
Rev Genet. 12:56–68. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sharma A, Costantini S and Colonna G: The
protein-protein interaction network of the human Sirtuin family.
Biochim Biophys Acta. 1834:1998–2009. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wu J, Tan Y, Deng H and Zhu D:
Relationship between degree-rank function and degree distribution
of protein-protein interaction networks. Comput Biol Chem. 32:1–4.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wu J, Tan YJ, Deng HZ and Zhu DZ: A new
measure of heterogeneity of complex networks based on degree
sequence. Unifying Themes in Complex Systems. 66–73. 2010.
View Article : Google Scholar
|
|
46
|
Dong J and Horvath S: Understanding
network concepts in modules. BMC Syst Biol. 1:242007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Freeman LC: Centrality in social networks
conceptual clarification. Soc Networks. 1:215–239. 1978. View Article : Google Scholar
|
|
48
|
Estrada E: Quantifying network
heterogeneity. Phys Rev E Stat Nonlin Soft Matter Phys.
82:0661022010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Nepusz T, Yu H and Paccanaro A: Detecting
overlapping protein complexes in protein-protein interaction
networks. Nat Methods. 9:471–472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Peng K, Vucetic S, Radivojac P, Brown CJ,
Dunker AK and Obradovic Z: Optimizing long intrinsic disorder
predictors with protein evolutionary information. J Bioinform
Comput Biol. 3:35–60. 2015. View Article : Google Scholar
|
|
51
|
He XX, Chang Y, Meng FY, Wang MY, Xie QH,
Tang F, Li PY, Song YH and Lin JS: MicroRNA-375 targets AEG-1 in
hepatocellular carcinoma and suppresses liver cancer cell growth in
vitro and in vivo. Oncogene. 31:3357–3369. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
3:e2642005. View Article : Google Scholar
|
|
55
|
Costantini S, Di Bernardo G, Cammarota M,
Castello G and Colonna G: Gene expression signature of human HepG2
cell line. Gene. 518:335–345. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Nagase T, Miyajima N, Tanaka A, Sazuka T,
Seki N, Sato S, Tabata S, Ishikawa K, Kawarabayasi Y, Kotani H, et
al: Prediction of the coding sequences of unidentified human genes.
III. The coding sequences of 40 new genes (KIAA0081-KIAA0120)
deduced by analysis of cDNA clones from human cell line KG-1. DNA
Res. 2:37–43. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Charrasse S, Mazel M, Taviaux S, Berta P,
Chow T and Larroque C: Characterization of the cDNA and pattern of
expression of a new gene over-expressed in human hepatomas and
colonic tumors. Eur J Biochem. 234:406–413. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Takeshita N, Mania D, Herrero S, Ishitsuka
Y, Nienhaus GU, Podolski M, Howard J and Fischer R: The cell-end
marker TeaA and the microtubule polymerase AlpA contribute to
microtubule guidance at the hyphal tip cortex of Aspergillus
nidulans to provide polarity maintenance. J Cell Sci.
126:5400–5411. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wu Z, Chen Y, Yang T, Gao Q, Yuan M and Ma
L: Targeted ubiquitination and degradation of G-protein-coupled
receptor kinase 5 by the DDB1-CUL4 ubiquitin ligase complex. PLoS
One. 7:e439972012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Golsteyn RM, Mundt KE, Fry AM and Nigg EA:
Cell cycle regulation of the activity and subcellular localization
of Plk1, a human protein kinase implicated in mitotic spindle
function. J Cell Biol. 129:1617–1628. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li JZ, Bunney BG, Meng F, Hagenauer MH,
Walsh DM, Vawter MP, Evans SJ, Choudary PV, Cartagena P, Barchas
JD, et al: Circadian patterns of gene expression in the human brain
and disruption in major depressive disorder. Proc Natl Acad Sci
USA. 110:pp. 9950–9955. 2013, View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Vinciguerra M, Mazzoccoli G, Piccoli C,
Tataranni T, Andriulli A and Pazienza V: Exploitation of host clock
gene machinery by hepatitis viruses B and C. World J Gastroenterol.
19:8902–8909. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Masri S, Papagiannakopoulos T, Kinouchi K,
Liu Y, Cervantes M, Baldi P, Jacks T and Sassone-Corsi P: Lung
adenocarcinoma distally rewires hepatic circadian homeostasis.
Cell. 165:896–909. 2016. View Article : Google Scholar : PubMed/NCBI
|