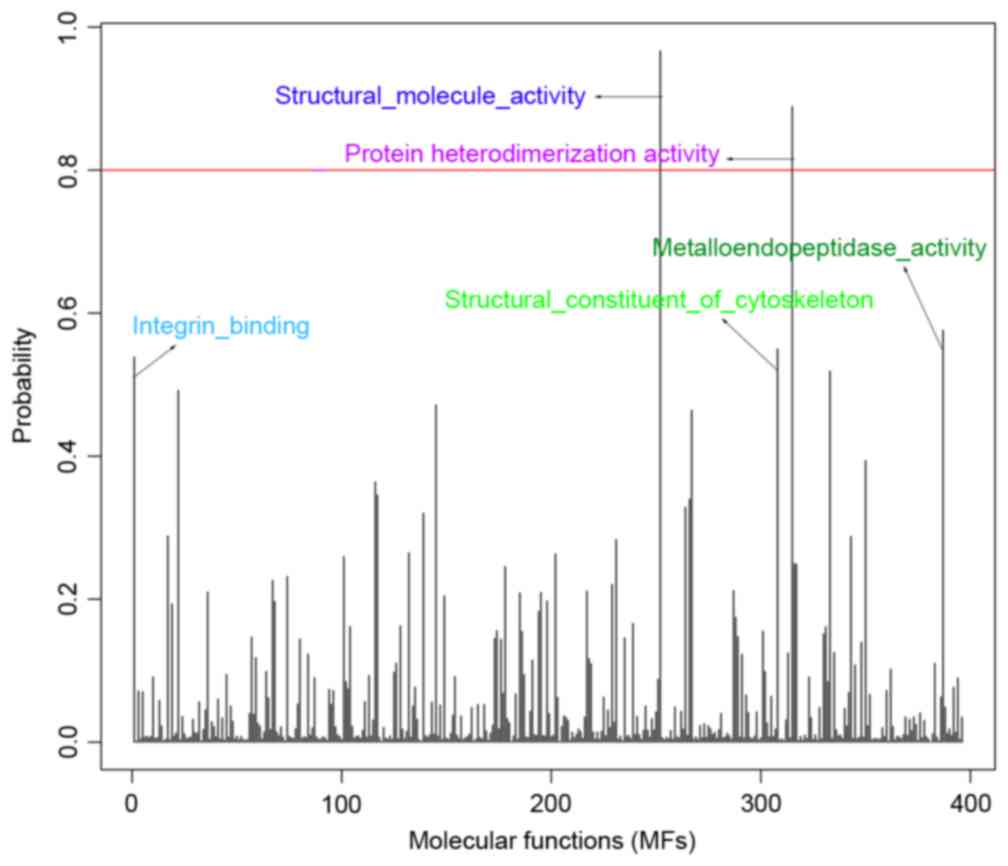
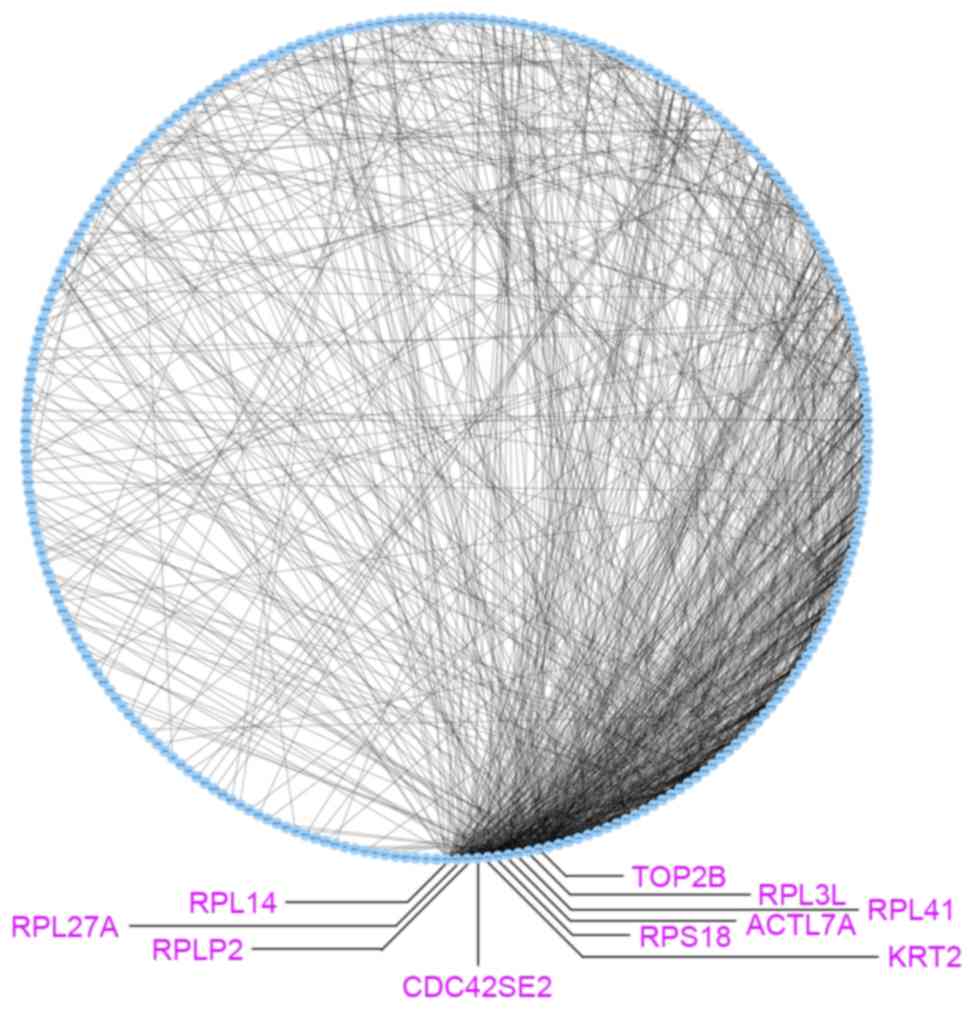
|
1
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Müller A, Homey B, Soto H, Ge N, Catron D,
Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al:
Involvement of chemokine receptors in breast cancer metastasis.
Nature. 410:50–56. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
DeSantis C, Siegel R, Bandi P and Jemal A:
Breast cancer statistics, 2011. CA Cancer J Clin. 61:409–418. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
DiSipio T, Rye S, Newman B and Hayes S:
Incidence of unilateral arm lymphoedema after breast cancer: A
systematic review and meta-analysis. Lancet Oncol. 14:500–515.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Charif M, Lower EE, Kennedy D, Kumar H,
Khan S, Radhakrishnan N and Zhang X: Abstract P3-05-16: The effect
of HER-2/neu inhibition on prolonging clinical benefit with
fulvestrant treatment for metastatic estrogen receptor positive
breast cancer patients treated with trastuzumab. Cancer Res.
75:3–5. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mittendorf EA, Clifton GT, Holmes JP,
Clive KS, Patil R, Benavides LC, Gates JD, Sears AK, Stojadinovic
A, Ponniah S and Peoples GE: Clinical trial results of the
HER-2/neu (E75) vaccine to prevent breast cancer recurrence in
high-risk patients: From US Military Cancer Institute Clinical
Trials Group Study I-01 and I-02. Cancer. 118:2594–2602. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Holder AM, Gonzalez-Angulo AM, Chen H,
Akcakanat A, Do KA, Symmans Fraser W, Pusztai L, Hortobagyi GN,
Mills GB and Meric-Bernstam F: Increased stearoyl-CoA desaturase 1
expression is associated with shorter survival in breast cancer
patients. Cancer Res. 72:6822012. View Article : Google Scholar
|
|
8
|
Gene Ontology Consortium, . Blake JA,
Dolan M, Drabkin H, Hill DP, Li N, Sitnikov D, Bridges S, Burgess
S, Buza T, et al: Gene Ontology annotations and resources. Nucleic
Acids Res. 41:(Database issue). D530–D535. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen DT, Nasir A, Culhane A, Venkataramu
C, Fulp W, Rubio R, Wang T, Agrawal D, McCarthy SM, Gruidl M, et
al: Proliferative genes dominate malignancy-risk gene signature in
histologically-normal breast tissue. Breast Cancer Res Treat.
119:335–346. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of Affymetrix GeneChip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bolstad B: Affy: Built-in Processing
Methods. 2013.
|
|
14
|
Lee J and Kim DW: Efficient multivariate
feature filter using conditional mutual information. Electronics
Lett. 48:161–162. 2012. View Article : Google Scholar
|
|
15
|
Quiroz-Zárate A, Haibe-Kains B and
Quackenbush J: BAGS: A Bayesian Approach for Geneset Selection.
2013.
|
|
16
|
Rasnake N, Haddad L, Rasnake M and Heidel
E: 487: Bacterial colonization of manual resuscitation bags. Crit
Care Med. 40:1–328. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kozumi H and Kobayashi G: Gibbs sampling
methods for Bayesian quantile regression. J Statistical Computa
Simulation. 81:1565–1578. 2011. View Article : Google Scholar
|
|
18
|
El-Hay T, Friedman N and Kupferman R:
Gibbs sampling in factorized continuous-time Markov processes.
arXiv preprint arXiv. 1206:32512012.
|
|
19
|
Chib S and Winkelmann R: Markov chain
Monte Carlo analysis of correlated count data. J Bus Eco
Statistics. 19:428–435. 2012. View Article : Google Scholar
|
|
20
|
Sinclair A: Algorithms for random
generation and counting: A Markov chain approach. Springer Sci Bus
Media. 2012.
|
|
21
|
Moradkhani H, DeChant CM and Sorooshian S:
Evolution of ensemble data assimilation for uncertainty
quantification using the particle filter-Markov chain Monte Carlo
method. Water Resources Res. 48:2012. View Article : Google Scholar
|
|
22
|
Gonzalez J, Low Y, Gretton A and Guestrin
C: Parallel gibbs sampling: From colored fields to thin junction
trees. International Conference on Artificial Intelligence and
Statistics. pp. 324–332. 2011
|
|
23
|
Yan Y and Zhang S: An improved estimation
method and empirical properties of the probability of informed
trading. J Banking Finance. 36:454–467. 2012. View Article : Google Scholar
|
|
24
|
Dawson JA, Ye S and Kendziorski C:
R/EBcoexpress: An empirical Bayesian framework for discovering
differential co-expression. Bioinformatics. 28:1939–1940. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Moon TK: The expectation-maximization
algorithm. Signal Pro Magazine IEEE. 13:47–60. 1996. View Article : Google Scholar
|
|
26
|
Haythornthwaite C: Social network
analysis: An approach and technique for the study of information
exchange. Library Inf Sci Res. 18:323–342. 1996. View Article : Google Scholar
|
|
27
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Grossglauser M and Vetterli M: Locating
nodes with EASE: Last encounter routing in ad hoc networks through
mobility diffusion. In: INFOCOM 2003. Twenty-Second Annual Joint
Conference of the IEEE Computer and Communications. IEEE Societies
IEEE. 1954–1964. 2003.
|
|
29
|
Hsiao TH, Chen HI, Roessler S, Wang XW and
Chen Y: Identification of genomic functional hotspots with copy
number alteration in liver cancer. EURASIP J Bioinform Syst Biol.
2013:142013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hong WX, Huang A, Lin S, Yang X, Yang L,
Zhou L, Huang H, Wu D, Huang X, Xu H and Liu J: Differential
expression profile of membrane proteins in L-02 cells exposed to
trichloroethylene. Toxicol Ind Health. 32:1774–1783. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hao X, Sun B, Hu L, Lähdesmäki H, Dunmire
V, Feng Y, Zhang SW, Wang H, Wu C, Wang H, et al: Differential gene
and protein expression in primary breast malignancies and their
lymph node metastases as revealed by combined cDNA microarray and
tissue microarray analysis. Cancer. 100:1110–1122. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cizkova M, Cizeron-Clairac G, Vacher S,
Susini A, Andrieu C, Lidereau R and Bièche I: Gene expression
profiling reveals new aspects of PIK3CA mutation in
ERalpha-positive breast cancer: Major implication of the Wnt
signaling pathway. PLoS One. 5:e156472010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lan J, Huang HY, Lee SW, Chen TJ, Tai HC,
Hsu HP, Chang KY and Li CF: TOP2A overexpression as a poor
prognostic factor in patients with nasopharyngeal carcinoma. Tumor
Biol. 35:179–187. 2014. View Article : Google Scholar
|
|
34
|
Ogawa R, Ishiguro H, Kuwabara Y, Kimura M,
Mitsui A, Mori Y, Mori R, Tomoda K, Katada T, Harada K and Fujii Y:
Identification of candidate genes involved in the radiosensitivity
of esophageal cancer cells by microarray analysis. Dis Esophagus.
21:288–297. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang L, Huang J, Jiang M and Sun L:
Survivin (BIRC5) cell cycle computational network in human no-tumor
hepatitis/cirrhosis and hepatocellular carcinoma transformation. J
Cell Biochem. 112:1286–1294. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Holbro T, Beerli RR, Maurer F, Koziczak M,
Barbas CF III and Hynes NE: The ErbB2/ErbB3 heterodimer functions
as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor
cell proliferation. Proc Natl Acad Sci USA. 100:pp. 8933–8938.
2003, View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Emde A, Köstler WJ and Yarden Y:
Association of Radiotherapy and Oncology of the Mediterranean arEa
(AROME): Therapeutic strategies and mechanisms of tumorigenesis of
HER2-overexpressing breast cancer. Crit Rev Oncol Hematol. 84 Suppl
1:e49–e57. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Montanaro L, Treré D and Derenzini M:
Changes in ribosome biogenesis may induce cancer by down-regulating
the cell tumor suppressor potential. Biochim Biophys Acta.
1825:101–110. 2012.PubMed/NCBI
|