Introduction
Nasopharyngeal carcinoma is a malignant tumor of
nasopharyngeal mucosa with high incidence in Guangdong, Guangxi and
other regions in China (1).
Epstein-Barr virus infection is closely associated with the
carcinogenesis of nasopharyngeal carcinoma. Nasopharyngeal cancer
is highly malignant, has distant metastasis in the early stages,
and is mainly located in cervical lymph nodes (2). Epithelial-mesenchymal transition (EMT)
refers to the transformation of epithelial cells into motile
mesenchymal cells, which is an important biological process for
epithelial cell-derived malignant tumor cells to obtain migration
and invasion capabilities. After EMT, cell morphology is altered,
with increased and thickened cell surface fibers and increased
pseudopodia. The expression of epithelial cell markers E-cadherin
and β-catenin are decreased, whereas the expression of mesenchymal
cell markers fibronectin, N-cadherin and vimentin are increased,
resulting in the increase of cell migration capacity and tumor
metastasis (3–5). Following EMT, epithelial cells in a
static state change into mesenchymal cells with a strong migration
ability. Moreover, proteolytic enzymes, such as matrix
metalloproteinase-9 (MMP-9), can degrade the basement membrane,
thereby facilitating cells to invade the extracellular matrix
(6). During the process of EMT in
nasopharyngeal carcinoma cells, these markers have similar changes,
but the mechanism leading to these changes remains unclear.
miRNAs affect the cell apoptosis, proliferation and
differentiation processes by regulating the expression of target
genes, and they are probably associated with tumor metastasis
(7). Studies have reported that
miRNAs are involved in the carcinogenesis and development of
nasopharyngeal carcinoma (8–10). At present, changes of 35 kinds of
miRNA expression levels have been found in nasopharyngeal carcinoma
tissue (11). The mutual effect of
miR141 and tumor-associated genes c-myc and PTEN promotes the
carcinogenesis and development of tumors (12). MicroRNA microarray analysis has shown
there is a significant difference between miR-10b expression in
nasopharyngeal carcinoma cells and normal nasopharyngeal epithelial
cells (13). To evaluate the role of
miR-10b in the carcinogenesis and development of nasopharyngeal
carcinoma, we used lentivirus to infect normal nasopharyngeal
epithelial cells aiming to observe cell proliferation and migration
changes, and to analyze the difference of expression levels in
epithelial cell and stromal cell markers.
Materials and methods
Cells
The nasopharyngeal carcinoma cell line CNE1, was
stored in our laboratory and cultured in RPMI-1640 medium
containing 10% calf serum (100 U/ml penicillin and 100 µg/ml
streptomycin). The immortalized nasopharyngeal epithelial cell line
NP69, was cultured with the same RPMI-1640 medium to which growth
factors were added. The cells were cultured at 37°C in a 5%
CO2 incubator.
Quantitative PCR
Total RNA was extracted using the TRIzol kit
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
RT-PCR was performed according to the manufacturer's instructions.
The reverse transcription conditions were as follows: 25°C for 5
min, 42°C for 30 min, 85°C for 5 min to inactivate the RNA enzyme.
qPCR was performed with U6 snRNA as the internal reference, and the
reaction conditions were: Pre-denaturation at 95°C for 30 sec,
denaturation at 95°C for 5 sec, followed by 40 cycles of annealing
at 60°C for 30 sec and extension at 72°C for 30 sec. The Cq values
of miR-10b and U6 were calculated according to the amplification
curves. Cq was calculated using the formula: Cq =
CqmiR-10b-CqU6. The expression level of
miR-10b in CNE1 and NP69 cells was compared using the
2−Cq method.
NP69 cells infected with lentivirus-miR-10b. The
lentivirus system was constructed to express miR-10b. The sequence
of miR-10b was inserted into the lentiviral plasmid pLP-VSVG.
PLP-VSVG-miR-10b, pLP1 and pLP2 were co-transfected into 293 T
cells, and after 48 h, lentivirus-containing supernatant was
collected. NP69 cells (1×106) were seeded in 6-well
plates, cultured overnight into monolayer cells, and then incubated
with lentivirus supernatant for 24 h to allow the lentivirus to
infect cells. Lentivirus-infected cells were collected on the same
day and days 2–6 after infection and the expression levels of
miR-10b were detected by RT-qPCR. NP69 cells infected with blank
pLP-VSVG served as the control.
CCK-8 assay
Lentivirus-infected NP69 cells were seeded in
96-well plates with 5×103 cells/well, and cultured
overnight at 37°C in a 5% CO2 incubator. Then, 10 µl of
the cell counting kit-8 (CCK-8) solution was added to each well,
and incubated for 24 h. The absorbance at a wavelength of 450 nm
was measured. NP69 cells infected with the blank lentivirus served
as the control. Cell viability was calculated using the formula:
Cell viability = (the test well OD value - the blank well OD
value)/(the control well OD value - the blank well OD value) ×
100%.
Cell scratch assay
Lentiviral-infected NP69 cells were cultured in
24-well plates with 5×104 cells/well, and incubated at
37°C in a 5% CO2 incubator until the cell density
reached 90%. The cell monolayer was scraped in a straight line to
create a scratch with a 10 µl pipette tip, and any debris was
removed by washing the cells once with PBS and the cells were
cultured continuously. Scratches were observed under a microscope
(Olympus BX53; Olympus Corporation, Tokyo, Japan) at 24, 48 and 72
h, the size of the scratches was measured, and the results
represented the mean of the three experiments. CNE1 and NP69 cells
without lentivirus infection served as controls.
Western blot analysis
NP69 cells were collected at 72 h after lentiviral
infection and lysed with lysate. The lysate was separated by 10%
sodium dodecyl sulphate-polyacrylamide gel electrophoresis and then
transferred to a polyvinylidene fluoride membrane and blocked with
5% skim milk at room temperature for 2 h. The membrane was
incubated, using the following mouse anti human primary antibodies:
Anti-E-cadherin, anti-β-catenin, anti-fibronectin, anti-vimentin
and anti-MMP9 (dilution, 1:1,500; cat. nos. ab1416; ab22656;
ab6328; ab8978; ab58803 respectively, purchased from Abcam plc.,
Burlingame, CA, USA) at room temperature for 1 h. The membrane was
then incubated with HRP-conjugated rabbit anti-mouse secondary
polyclonal antibody (dilution, 1:2,500; cat. no. ab6728; Abcam
plc.) for 1 h. The HRP enzyme substrate was added and incubated for
5 min to develop color, and the immunoblotting result was recorded
using a fully automated western blot WEs analysis system
(ProteinSimple, San Jose, CA, USA). The density of each band was
determined by ImageJ software, and β-actin was used as the internal
reference. CNE1 and NP69 cells infected with blank lentivirus were
used as controls. The relative expression level was calculated as
the ratio of the assessed protein and β-actin and expressed as the
mean ± standard deviation (n=3).
Statistical analysis
Data were expressed as mean ± standard deviation,
the difference between two groups was compared with the Student's
t-test, and multiple group comparison was performed using Fisher's
LSD test. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-10b expression in cells
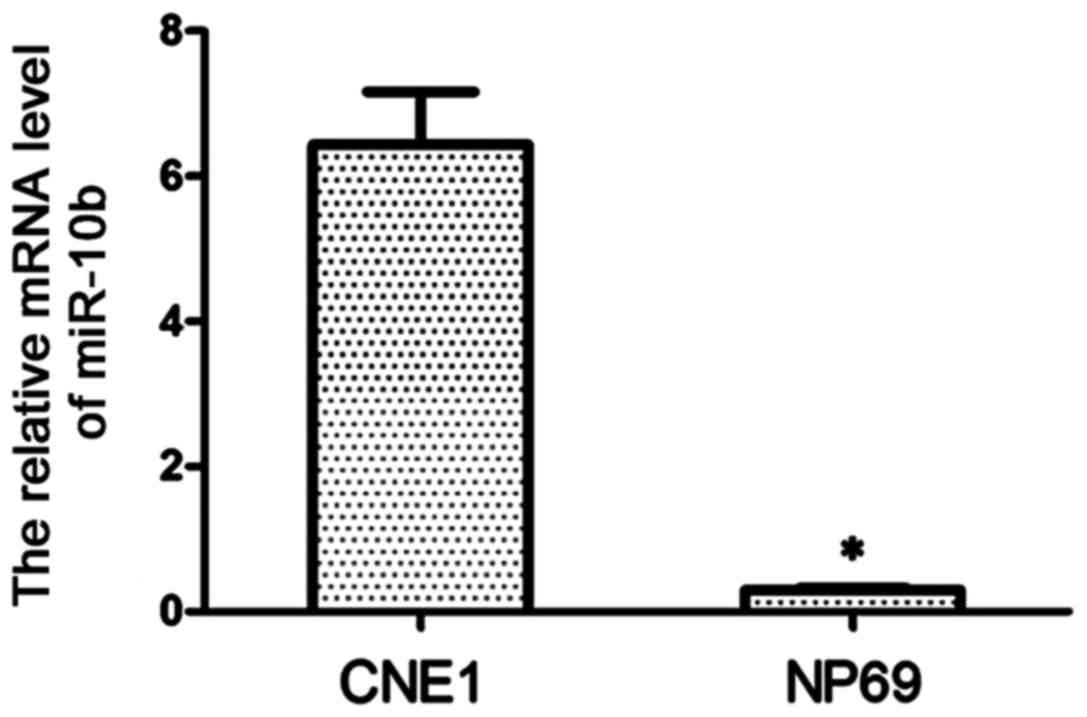
The expression of miR-10b in CNE1 cells was detected
by RT-qPCR. As shown in Fig. 1, the
expression level of miR-10b in CNE1 cells was significantly higher
than that in NP69 cells (P<0.01), and miR-10b was almost absent
in NP69 cells.
miR-10b expression in
lentivirus-infected NP69 cells
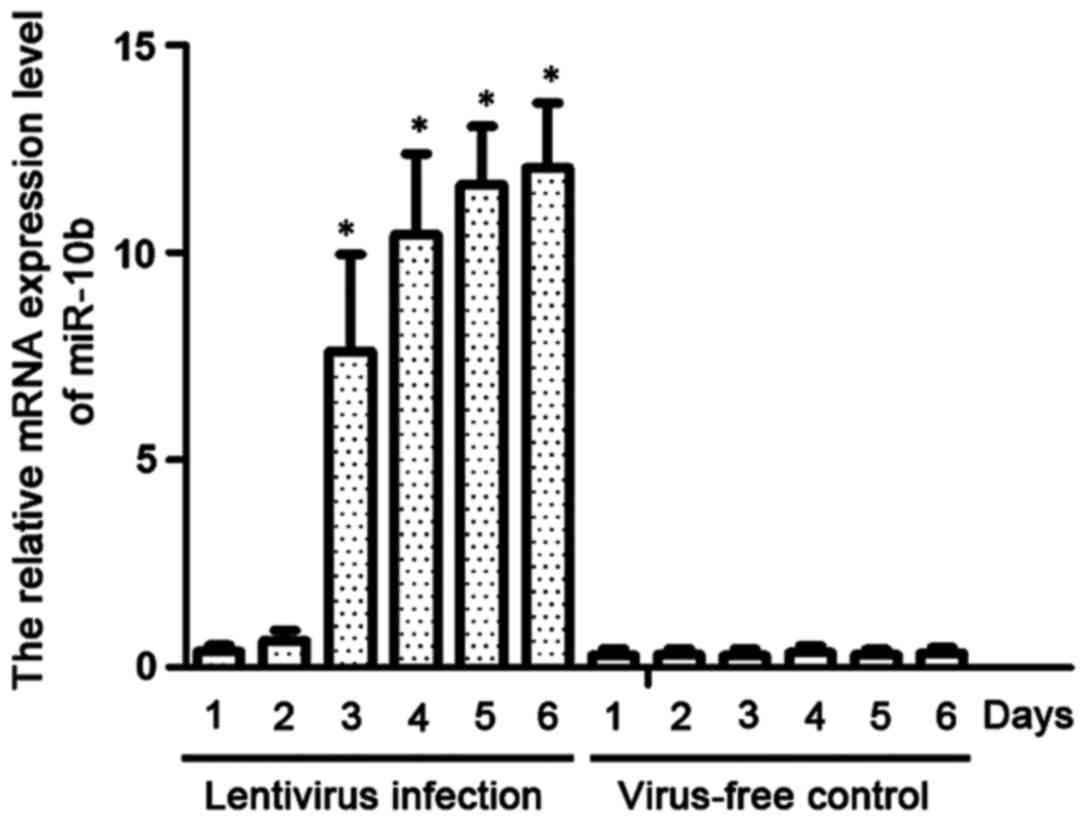
We established a cell line stably expressing miR-10b
by infecting NP69 cells with lentivirus. RT-qPCR was used to detect
the expression levels of miR-10b. As shown in Fig. 2, miR-10b was highly expressed on day 3
after lentiviral infection, and its level on day 5 was
significantly higher than that of NP69 cells with blank lentiviral
infection (P<0.01).
miR-10b promotes NP69 cell
proliferation
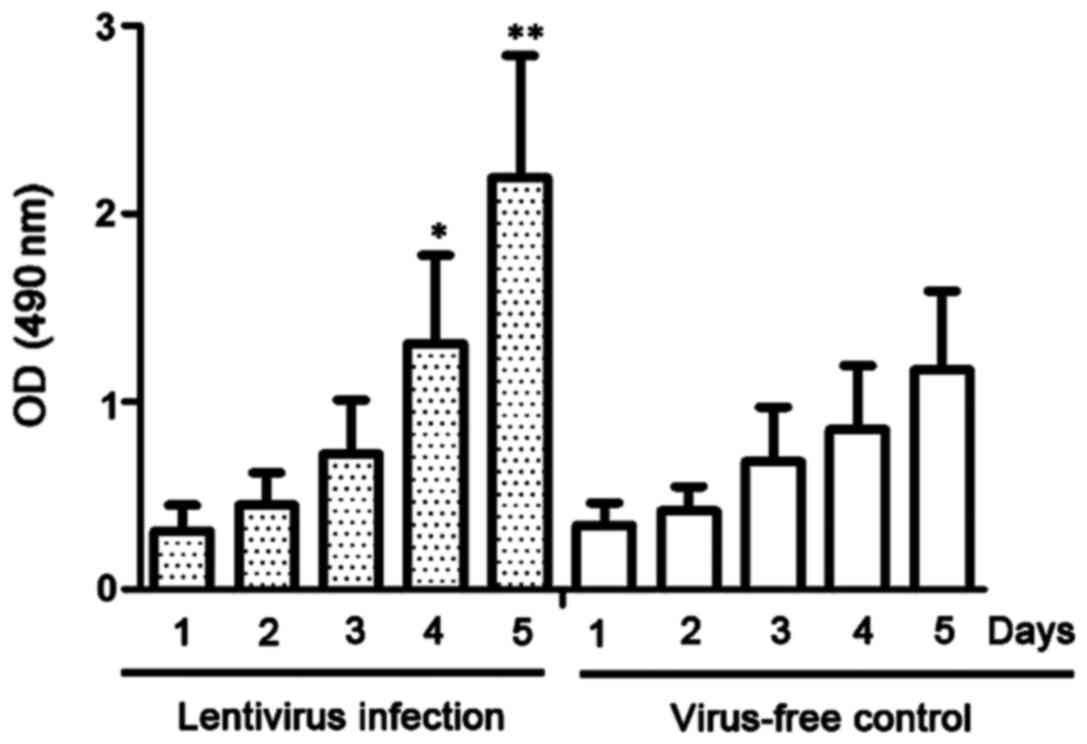
A CCK-8 assay was used to detect the proliferative
ability of lentivirus-infected NP69 cells. As shown in Fig. 3, the proliferation of NP69 cells
stably expressing miR-10b was significantly more rapid than that of
NP69 cells with no lentivirus infection at day 4 after infection
(P<0.05).
miR-10b promotes the migration of NP69
cells
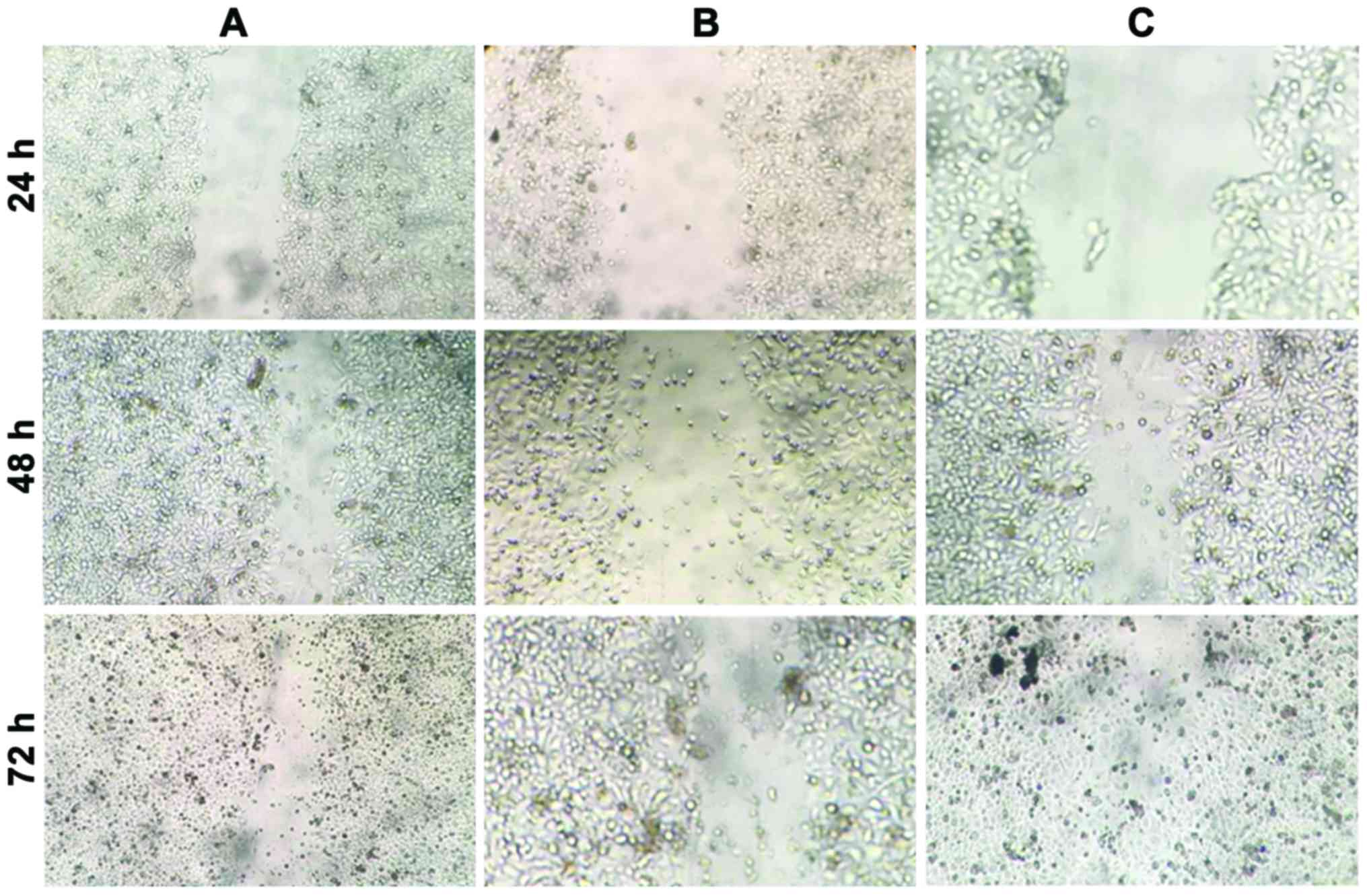
The effect of miR-10b on the migration of NP69 cells
was examined using a cell scratch assay. As shown in Fig. 4 and Table
I, following cell scratching, CNE1 and NP69 cells expressing
miR-10b grew and covered half of the scratches at 48 h, and almost
all the scratches after 72 h. By contrast, NP69 cells in the
control group did not cover half of the scratches after 72 h.
miR-10b promoted the migration of NP69 cells in a time-dependent
manner. The difference in expression levels of miR-10b between CNE1
and NP69 cells led to a difference in the cell migration capacity,
thereby further resulting in cancer cell metastases.
 | Table I.Cell scratch size (mean ± standard
deviation, n=3). |
Table I.
Cell scratch size (mean ± standard
deviation, n=3).
| Scratch size
(µm) | 24 h | 48 h | 72 h | P-value |
|---|
| NP69 cells stably
expressin miR-10b | 21.7±1.4 | 10.7±2.3 |
4.2±1.9 | <0.01 |
| NP69 cells infected
with blank lentivirus | 25.5±3.2 | 19.7±4.3 | 15.1±3.7 | <0.05 |
| CNE1 cells | 32.6±2.5 | 11.8±3.1 |
1.3±0.4 | <0.01 |
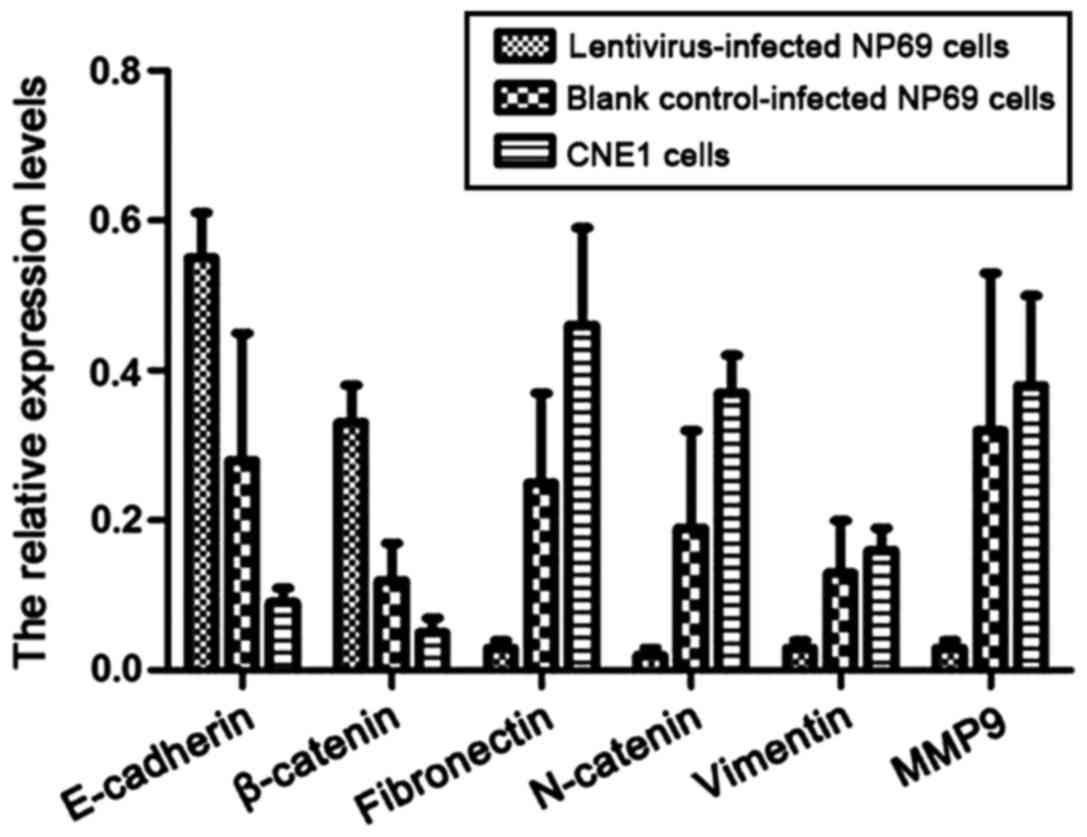
The expression levels of epithelial
cells and stromal cell markers
Western blot analysis was used to detect the
expression levels of epithelial cells and interstitial cell markers
in CNE1 cells and lentivirus-infected NP69 cells expressing
miR-10b. As shown in Fig. 5, the
expression levels of E-cadherin and β-catenin in CNE1 and NP69
cells stably expressing miR-10b were significantly decreased
compared to NP69 cells with no miR-10b expression (P<0.05). By
contrast, the expression levels of stromal cell markers,
fibronectin, N-catenin, vimentin and MMP-9 were significantly
increased (P<0.05).
Discussion
MicroRNAs regulate gene expression at
transcriptional levels, and increasing research has focused on the
role of microRNAs in the carcinogenesis and development of tumors.
Previous findings have shown there is a difference in terms of the
expression of miR-10b between nasopharyngeal carcinoma and normal
nasopharyngeal mucosa tissue (12).
The present findings showed that miR-10b expression in the CNE1
nasopharyngeal carcinoma cell line was higher than that in normal
nasopharyngeal mucosa. The expression of epithelial cell markers
E-cadherin and β-catenin, decreased after NP69 cells were infected
with lentivirus and expressed miR-10b. E-cadherin enhances
cell-cell adhesion, and its downregulation can decrease cell
adhesion and increase cell motility through the basement membrane
(14,15). The expression of stromal cell markers
fibronectin, N-cadherin, vimentin and MMP-9, was increased and the
cell migration ability was enhanced, indicating that miR-10b
promoted the EMT of nasopharyngeal carcinoma cells. miR-10b
promoted tumor cell invasion and migration by downregulating the
expression of KLF4 (16) and as a
direct target of miR-10b, KLF4 was capable of inhibiting tumor cell
invasion and migration (17), which
could be used for target for further study.
In summary, the present findings show that miR-10b
is involved in the EMT of nasopharyngeal carcinoma cells, promotes
cell proliferation and migration and is closely associated with the
metastasis of nasopharyngeal carcinoma. Further research is needed
to elucidate the molecular mechanism of miR-10b regulating the EMT
of nasopharyngeal carcinoma cells. miR-10b is expected to be a key
target in the treatment of nasopharyngeal carcinoma, providing new
opportunities for the development of new targeted therapies.
References
|
1
|
Spano JP, Busson P, Atlan D, Bourhis J,
Pignon JP, Esteban C and Armand JP: Nasopharyngeal carcinomas: An
update. Eur J Cancer. 39:2121–2135. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee AW, Poon YF, Foo W, Law SC, Cheung FK,
Chan DK, Tung SY, Thaw M and Ho JH: Retrospective analysis of 5037
patients with nasopharyngeal carcinoma treated during 1976–1985:
Overall survival and patterns of failure. Int J Radiat Oncol Biol
Phys. 23:261–270. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
He YX, Song XH, Zhao ZY and Zhao H: HOXA13
upregulation in gastric cancer is associated with enhanced cancer
cell invasion and epithelial-to-mesenchymal transition. Eur Rev Med
Pharmacol Sci. 21:258–265. 2017.PubMed/NCBI
|
|
4
|
Bu JQ and Chen F: TGF-β1 promotes cells
invasion and migration by inducing epithelial mesenchymal
transformation in oral squamous cell carcinoma. Eur Rev Med
Pharmacol Sci. 21:2137–2144. 2017.PubMed/NCBI
|
|
5
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chang JY, Wright JM and Svoboda KK: Signal
transduction pathways involved in epithelial-mesenchymal transition
in oral cancer compared with other cancers. Cells Tissues Organs.
185:40–47. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sarkar FH, Li Y, Wang Z, Kong D and Ali S:
Implication of microRNAs in drug resistance for designing novel
cancer therapy. Drug Resist Updat. 13:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang LJ, Chou YF, Chen PR, Su B, Hsu YC,
Chang CH and Lee JW: Differential miRNA expression in repeated
recurrence of nasopharyngeal carcinoma. Cancer Lett. 344:188–194.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Plieskatt JL, Rinaldi G, Feng Y, Levine
PH, Easley S, Martinez E, Hashmi S, Sadeghi N, Brindley PJ, Bethony
JM, et al: Methods and matrices: Approaches to identifying miRNAs
for nasopharyngeal carcinoma. J Transl Med. 12:32014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu N, Cui RX, Sun Y, Guo R, Mao YP, Tang
LL, Jiang W, Liu X, Cheng YK, He QM, et al: A four-miRNA signature
identified from genome-wide serum miRNA profiling predicts survival
in patients with nasopharyngeal carcinoma. Int J Cancer.
134:1359–1368. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen SJ, Chen GH, Chen YH, Liu CY, Chang
KP, Chang YS and Chen HC: Characterization of Epstein-Barr virus
miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One.
5:e127452010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang L, Deng T, Li X, Liu H, Zhou H, Ma
J, Wu M, Zhou M, Shen S, Li X, et al: microRNA-141 is involved in a
nasopharyngeal carcinoma-related genes network. Carcinogenesis.
31:559–566. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen HC, Chen GH, Chen YH, Liao WL, Liu
CY, Chang KP, Chang YS and Chen SJ: MicroRNA deregulation and
pathway alterations in nasopharyngeal carcinoma. Br J Cancer.
100:1002–1011. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sánchez-Tilló E, Lázaro A, Torrent R,
Cuatrecasas M, Vaquero EC, Castells A, Engel P and Postigo A: ZEB1
represses E-cadherin and induces an EMT by recruiting the SWI/SNF
chromatin-remodeling protein BRG1. Oncogene. 29:3490–3500. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huber GF, Züllig L, Soltermann A, Roessle
M, Graf N, Haerle SK, Studer G, Jochum W, Moch H and Stoeckli SJ:
Downregulation of E-Cadherin (ECAD) - a predictor for occult
metastatic disease in sentinel node biopsy of early squamous cell
carcinomas of the oral cavity and oropharynx. BMC Cancer. 11(217):
1–8. 2011.PubMed/NCBI
|
|
16
|
Xiao H, Li H, Yu G, Xiao W, Hu J, Tang K,
Zeng J, He W, Zeng G, Ye Z, et al: MicroRNA-10b promotes migration
and invasion through KLF4 and HOXD10 in human bladder cancer. Oncol
Rep. 31:1832–1838. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tian Y, Luo A, Cai Y, Su Q, Ding F, Chen H
and Liu Z: MicroRNA-10b promotes migration and invasion through
KLF4 in human esophageal cancer cell lines. J Biol Chem.
285:7986–7994. 2010. View Article : Google Scholar : PubMed/NCBI
|