Introduction
Oligodendroglial tumors, including
oligodendrogliomas (ODs) and oligoastrocytomas (OAs), present as
distinct histopathological and clinical subsets of glioma, with
slow growth and a favorable prognosis (1,2).
Oligodendroglial tumors account for <10% of the diffuse gliomas
(2). The histopathological diagnosis
of these tumors is difficult: OAs with typical morphological
features are histologically well defined, whereas other
non-classical tumors, OAs in particular, are more challenging to
diagnose due to equivocal definitions as a result of sharing
oligodendroglial and astrocytic morphology, as well as the
uncertainty of representative tumor sampling (2,3).
Therefore, histological diagnosis provides inadequate prognostic
information on oligodendroglial tumors, as demonstrated by markedly
varied survival times among patients with similar histological
features in the clinic (2).
Like other tumors, oligodendroglial tumors develop
as a result of heterogeneous genetic and molecular alterations
(1). These tumor-specific molecular
abnormalities determine the clinical and biological behavior of
each tumor; therefore certain molecular biomarkers may provide
useful clinical information (2). At
present, two identified biomarkers for oligodendroglial tumors are
mutations in isocitrate dehydrogenase (IDH)1 and
IDH2 (referred to collectively as IDH), and the loss
of heterozygosity of chromosome arms 1p and 19q (2). IDH mutations are observed in
70–80% of oligodendroglial tumors and are associated with an
improved survival time (2). The
complete co-deletion of 1p/19q resulting from an unbalanced
translocation is present in 60–80% of ODs, whereas it is observed
less frequency in OAs (10–15%) (4);
this genetic alteration is associated with a positive outcome and
response to chemotherapy (5). The
majority of 1p/19q co-deleted oligodendroglial tumors also exhibit
IDH mutations (6).
Sequencing studies have identified recurrent
mutations in the Capicua transcriptional repressor (CIC)
gene within 50–70% (2) of 1p/19q
co-deleted oligodendroglial tumors (1,3,4,7–10). CIC is situated on cytoband
19q13.2 and encodes a 164 kDa protein containing a high-mobility
group box (9). The gene was first
identified as a downstream repressor of the receptor tyrosine
kinase signaling pathway in regulating developmental patterning and
cell fate in Drosophila (11).
Mutations in this gene are almost exclusively observed in
oligodendroglial tumors (3,7). Certain mutations may result in
CIC repression; tumors with this mutation comprise an
aggressive subset of 1q/19q co-deleted gliomas (4). However, there are inconsistent reports
regarding the association between CIC mutations and prognosis in
patients with oligodendroglial tumors; some reported a significant
survival correlation of CIC mutations whiles others did not
(4,8,9).
Therefore, the aim of the present study was to investigate the
prognostic ability of altered CIC expression level as an
alternative to CIC mutation status among IDH-mutant
oligodendroglial tumors with or without co-deletion of 1p/19q.
Patients and methods
Patients and samples
Data and samples from patients with glioma who were
admitted between January 2003 and December 2012 to the Department
of Neurosurgery, Xijing hospital (Xi'an, China) were reviewed in
the present study. A total of 54 patients who met the following
inclusion criteria were included in the present study: i) Aged
>18 years; ii) with pathologically confirmed oligodendroglial
tumors, including ODs and OAs [World Health Organization (WHO)
grade II to III]; iii) carrying IDH1 mutations (as
determined by anti-IDH1R132H immunohistochemistry (IHC)
analysis at the Department of Pathology, Xijing hospital); iv) with
available records of 1p/19q status (determined by fluorescence
in situ hybridization analysis at the Department of
Pathology, Xijing hospital); and v) with written informed consent.
The histopathologic features and WHO grading were evaluated
according to the WHO 2007 classification (12). Formalin-fixed, paraffin-embedded tumor
tissues were examined in the present study. All patients underwent
the radical neurosurgery of primary tumors and received
stereotactic fractionated radiosurgery, with a median total dose of
36 Gy, postoperatively. Patient demographics and clinical data were
retrieved from the Xijing Hospital electronic medical record
system. Local institutional review board approval was obtained to
use archived material for study purposes. All procedures performed
with human tissue were in accordance with the ethical standards of
the institutional research committee and with the 1964 Helsinki
declaration and its later amendments, or comparable ethical
standards.
IHC staining for CIC protein
The protein expression of CIC was examined by
IHC staining. In the literature, the vast majority of mutations in
CIC occurred in exons 5 and 20 (3,9).
Therefore, an antibody specific for the C-terminus region of
CIC protein (cat. no. TA590520; OriGene Technologies, Inc.,
Rockville, MD, USA) was used. IHC staining was performed as
described previously (13); briefly,
paraffin-embedded sections were deparaffinized and rehydrated, and
antigen retrieval was performed. Subsequently the tumor sections
were immersed in methanol containing 3.0% hydrogen peroxide for 20
min to block endogenous peroxidase activity, and incubated in 2.5%
goat serum (Abcam, Cambridge, UK) to decrease non-specific binding.
Thereafter, tumor sections were incubated at 4°C overnight with the
primary anti-human CIC antibody (dilution, 1:1,000) followed by
incubation with a horseradish peroxidase-conjugated goat
anti-rabbit IgG antibody (dilution, 1:1,200; cat. no. A32732;
Pierce; Thermo Fisher Scientific, Inc., Waltham, MA, USA) for 30
min. Immunolabeled sections were visualized by staining with
3,3′-diaminobenzidine (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) and counterstaining with hematoxylin. Negative controls
were sections acquired from the same tumor without primary antibody
incubation.
The CIC nuclear staining pattern was evaluated in a
semiquantitative manner, as previously described (9). Briefly, the protein levels were scored
by multiplying the degree of intensity by percentage of stained
cells, with the intensity being graded as 0 (no staining), 1
(weak), 2 (moderate) and 3 (strong), and the percentage of stained
cells being classified as 0 (<10%), 1 (10–50%) and 2 (>50%).
The CIC scoring was performed by two independent investigators,
with any scoring disputes resolved through discussion.
Investigators were blinded to the clinical data of the assessed
sections, but not to the objectives of the study. The scores were
designated as absent (0), weak (1 and 2), moderate (3 and 4) or
strong (6).
Public datasets of oligodendroglial
tumors
The cancer genome atlas (TCGA)
Clinical and demographical data from 265 patients
(≥18 years old) with oligodendroglial tumors carrying IDH
mutations were retrieved from the TCGA data portal (https://tcga-data.nci.nih.gov/). Somatic
mutations in IDH and CIC were determined by level 2
Illumina Genome Analyzer DNA Sequencing data. For the assessment of
the 1p/19q status, level 3 Affymetrix Genome-Wide Human SNP 6.0
array data were analyzed within the GISTIC2.0 module at GenePattern
(http://genepattern.broadinstitute.org/gp). An
amplitude threshold of ± 0.2 was used. Additionally, CIC
mRNA expression levels were determined using level 3
IlluminaHiSeq_RNASeqV2 data.
Gene expression omnibus (GEO)
Data from a total of 45 patients (≥18 years old)
with oligodendroglial tumors carrying an IDH1 (R132)
mutation or the glioma-CpG island methylator phenotype (G-CIMP)
were obtained from the GEO data portal (http://www.ncbi.nlm.nih.gov/geo/) from the GSE16011
(n=30) (14) and GSE43388 (n=15)
(15) datasets. The 1p/19q status was
available for all samples. For each sample, CIC mRNA
expression levels were determined using the
log2-transformed RMA-normalized signal intensity from
the Affymetrix Human Genome Plus 2.0 Microarray platform. In
addition, expression profiles for 8 non-tumor brain tissues from
the GSE16011 dataset were downloaded as controls.
Cross-dataset/platform mRNA data processing
To account for differences in systematic measurement
between the distinct datasets and platforms, each set of CIC
mRNA expression data was independently standardized by transforming
the expression data to a mean of 0 and a standard deviation of 1,
as per a previously described method (16).
Gene set enrichment analysis (GSEA)
To assess the enrichment of gene sets within
different subgroups, GSEA was performed with 1,000 permutations, as
previously described (17). Gene sets
derived from the Biological Process Ontology (Molecular Signatures
Database; http://software.broadinstitute.org/gsea/msigdb) were
evaluated.
Statistical analysis
Overall survival (OS) time was defined as the time
from diagnosis until mortality or the final follow-up. Survival
data were analyzed using the Kaplan-Meier method and compared using
the log-rank test. Univariate Cox regression analysis was performed
to investigate the association between variables and OS time. A
multiple Cox regression model was applied to investigate the
independence of each prognostic indicator that was statistically
significant within univariate Cox models. The associations between
clinical and molecular variables were evaluated using a
χ2 test or Fisher's exact test. Differences in CIC
expression between groups were assessed by a Student's t-test or
Mann-Whitney U-test. The optimal threshold to separate patients
into the poor or good prognostic subgroups was determined by the
maximally selected rank statistics (18). All calculations were performed with
SPSS software (version 19.0; IBM Corporation, New York, NY, USA)
and R software (version 3.2.5; https://www.r-project.org/). P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression pattern of CIC among
IDH-mutant oligodendroglial tumors
The demographical and molecular characteristics of
all included patients are presented in Table I.
 | Table I.Characteristics of patients with
IDH-mutant oligodendroglial tumors. |
Table I.
Characteristics of patients with
IDH-mutant oligodendroglial tumors.
| Variables | Xijing | TCGA | GEO |
|---|
| Sample size, n | 54 | 265 | 45 |
| World Health |
|
|
|
| Organization grade,
n |
|
|
|
| II | 22 | 163 | 6 |
|
III | 32 | 102 | 39 |
| Tumor type, n |
|
|
|
|
OAs | 23 | 110 | 12 |
|
ODs | 31 | 155 | 33 |
| Sex, n |
|
|
|
|
Male | 29 | 143 | 29 |
|
Female | 25 | 122 | 16 |
| Age, years |
|
|
|
| Median
(range) | 43 | 41 | 45 |
|
| (18–69) | (18–74) | (28–75) |
| <50,
n | 35 | 184 | 27 |
| ≥50,
n | 19 | 81 | 18 |
| Karnofsky
performance status |
|
|
|
| Median
(range) | 80 | 90 | 100 |
|
| (50–100) | (60–100) | (10–100) |
| <80,
n | 21 | 3 | 8 |
| ≥80,
n | 28 | 59 | 26 |
| IDH mutation
type, n |
|
|
|
|
IDH1 | 54 | 247 |
45a |
|
IDH2 | 0 | 18 | – |
| 1p/19q status,
n |
|
|
|
|
Co-deletion | 28 | 153 | 29 |
|
Intact | 26 | 112 | 16 |
| CIC mutation
status, n |
|
|
|
|
CIC mutant | – | 104 | – |
|
CIC wide-type | – | 161 | – |
By comparing CIC mRNA expression between
tumor and non-tumor brain tissue from TGCA, it was identified that
CIC mutations were associated with decreased CIC mRNA
expression levels (unpaired t-test, P<0.001; data not shown),
and were almost exclusively observed in tumors with 1p/19q
co-deletion (Fisher's exact test, P<0.001; data not shown).
Additionally, CIC mRNA expression was increased in patients
aged <50 years compared with those ≥50, but no significant
difference was observed between CIC mRNA expression and sex
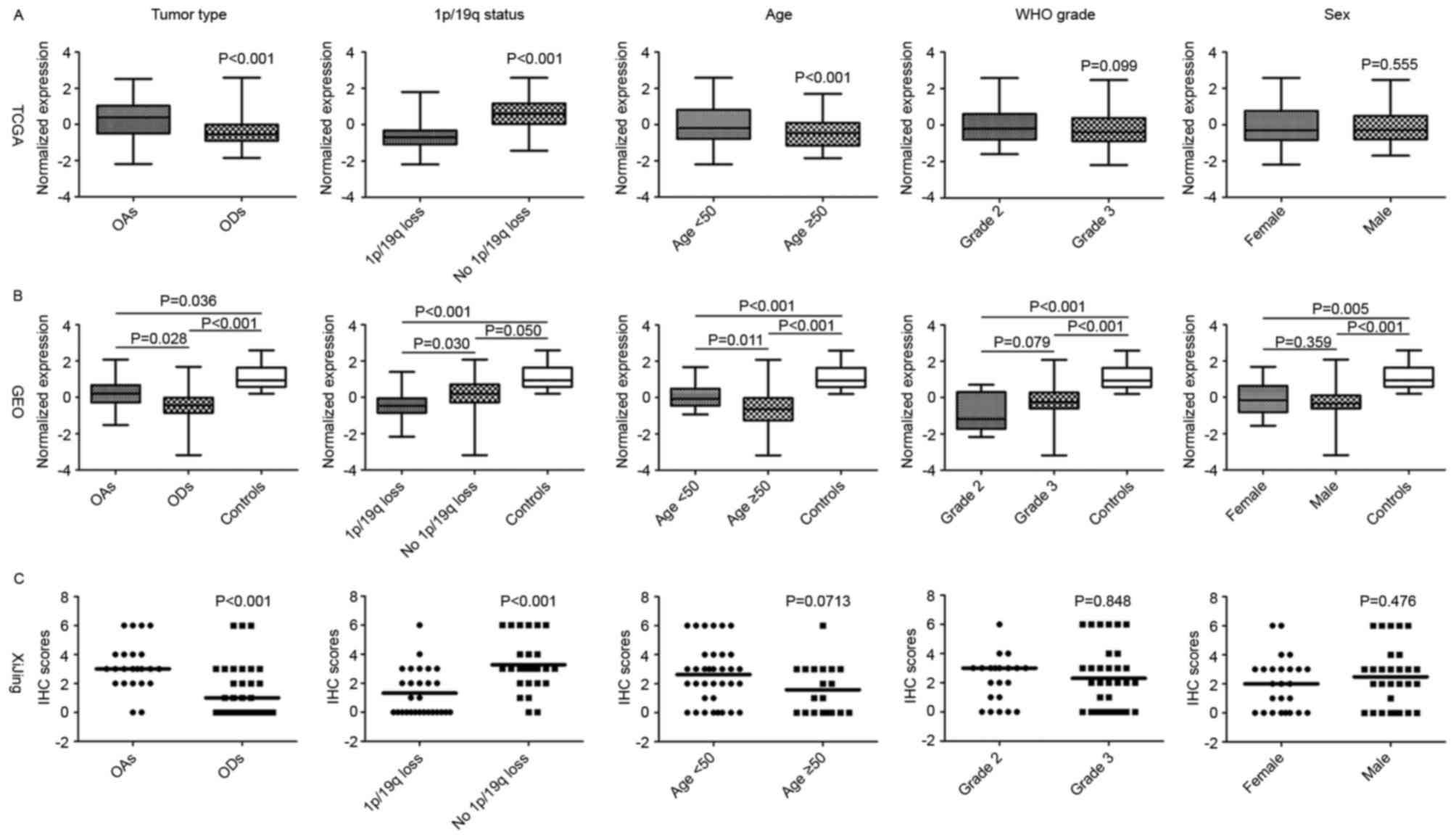
(Fig. 1A).
In the GEO samples, CIC expression was
identified as significantly repressed in IDH-mutant
oligodendroglial tumors relative to non-tumor tissue (unpaired
t-test, P<0.05). Among tumors with distinct clinical or
molecular features, OAs were associated with increased CIC
mRNA expression compared with ODs, whereas no significant
differences in CIC expression between grade II and III
tumors were identified. Regarding 1p/19q status, CIC mRNA
expression was significantly decreased in tumors with 1p/19q
co-deletion compared with those without (Fig. 1B).
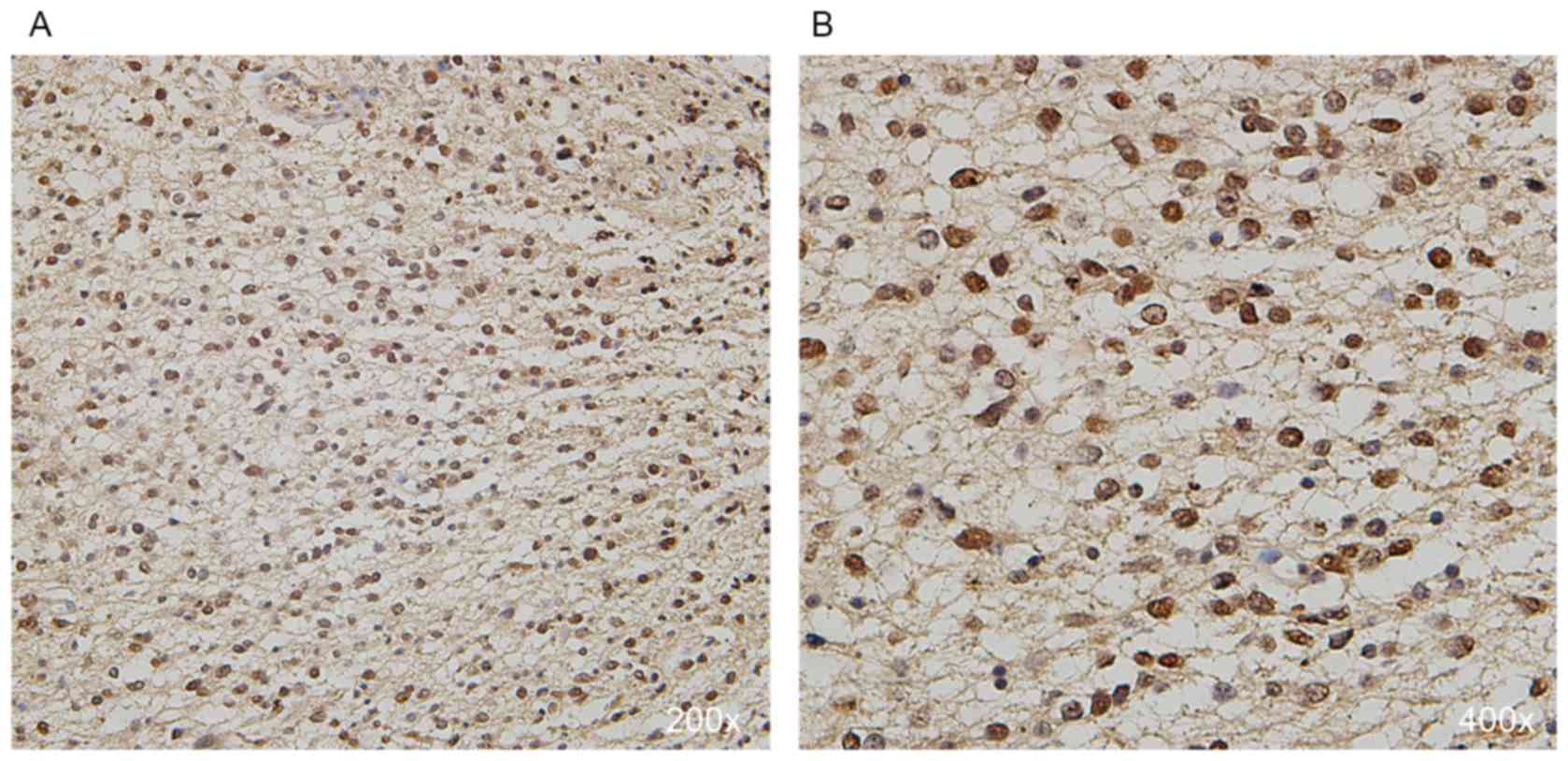
In the IHC examination of the tumor samples, it was
observed that 16 (30%), 13 (24%), 18 (33%) and 7 (13%) cases were
detected with absent, weak, moderate and strong nuclear expression
of CIC, respectively (Fig. 2). All 4
cases with discrepancy were defined as weakly (n=3) or moderately
(n=1) stained tumors after discussion (Three tumors were initially
defined as moderately stained by one investigator but weakly
stained by the other, while one tumor was initially defined as
strongly stained by one investigator but moderately stained by the
other). The protein expression patterns were consistent with the
mRNA levels from the GEO and TGCA data (Fig. 1). Notably, absent CIC expression was
significantly associated with 1p/19q co-deleted tumors
(χ2 test, P<0.001) and ODs (χ2 test,
P=0.006), whereas strong expression was associated with tumors
without 1p/19q co-deletion (χ2 test, P=0.047; Fig. 1C).
Increased CIC expression is associated
with a favorable prognosis in IDH-mutant, 1p/19q co-deleted
oligodendroglial tumors
Of the 153 oligodendroglial tumors from TCGA with an
IDH mutation and 1p/19q co-deletion, CIC mutations
were detected in 100 (65.4%) cases. No significant association was
identified between CIC mutations and OS time among TCGA
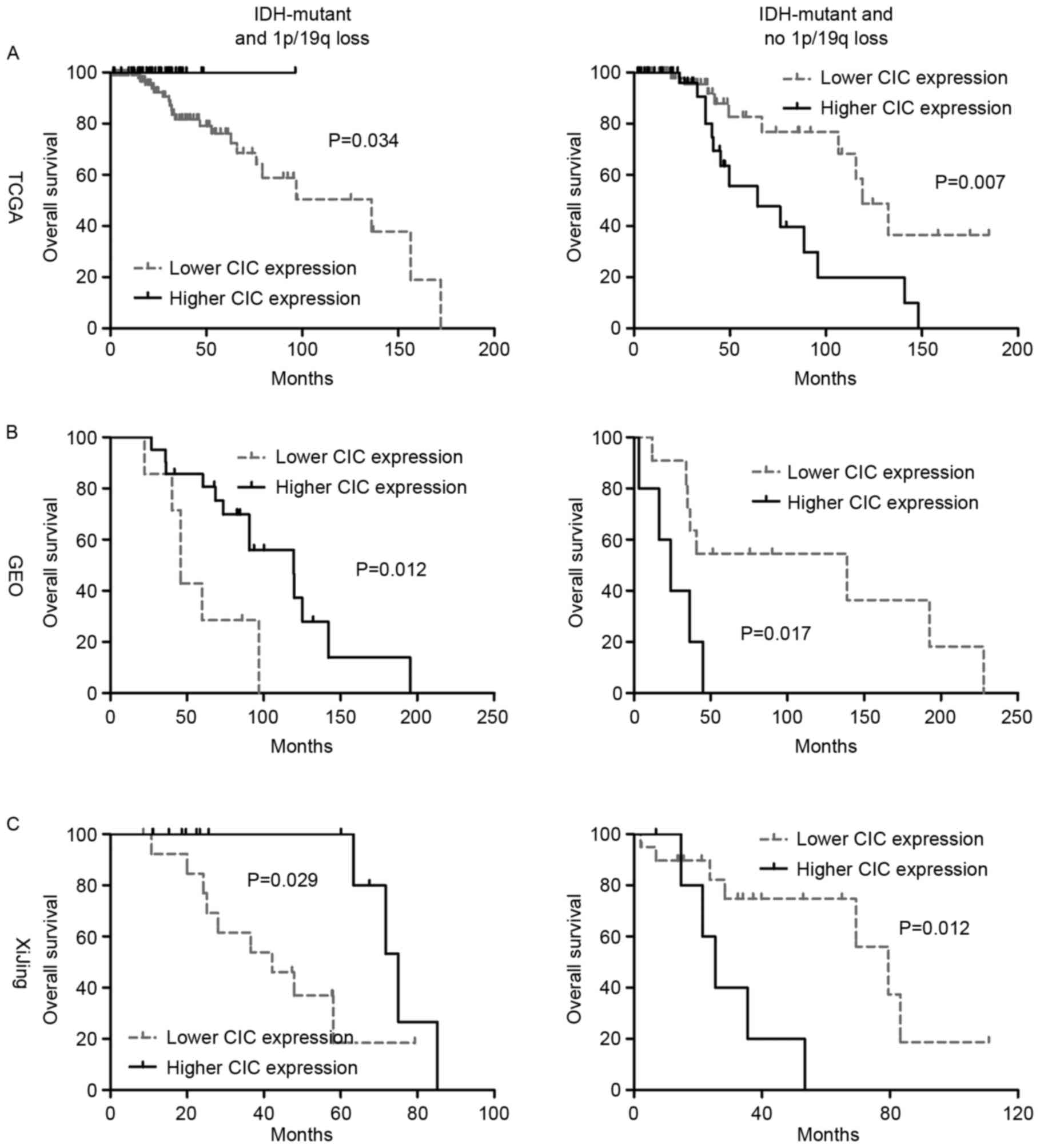
samples (log-rank P=0.225). However, increased CIC expression was
associated with a favorable prognosis in IDH-mutant
oligodendroglial tumors with 1p/19q co-deletion (median OS time,
higher vs. lower, not reached vs. 136.1 months; log-rank P=0.034;
Fig. 3A). Similarly, among
IDH-mutant and 1p/19q co-deleted oligodendroglial tumors
from GEO, higher CIC expression was associated with longer OS time
compared with lower CIC expression (median OS time, higher vs.
lower, 119.5 vs. 45.7 months; log-rank P=0.012; Fig. 3B). Additionally, in the IHC
examination of the present study cohort, patients with absent CIC
expression exhibited a significantly shorter OS than those with
detectable protein expression (weak, moderate or strong; median OS,
absent vs. detectable, 75.0 vs. 42.2 months; log-rank P=0.029;
Fig. 3C). These data collectively
demonstrate that higher CIC expression is a favorable indicator for
OS time among patients with oligodendroglial tumors with IDH
mutation and 1p/19q co-deletion.
Higher CIC expression is associated
with poor prognosis in IDH-mutant oligodendroglial tumors without
the 1p/19q co-deletion
In contrast to IDH-mutant, 1p/19q co-deleted
oligodendroglial tumors, only 4 cases (3.6%) were detected with
CIC mutations in those without 1p/19q co-deletion from TCGA.
In this subset of oligodendroglial tumors, higher CIC expression
was associated with poorer OS (median OS, higher vs. lower, 64.4
vs. 119.0 months; log-rank P=0.007; Fig.
3A). The distinct prognostic ability was also confirmed in GEO
samples (median OS, higher vs. lower, 23.8 vs. 138.8 months;
log-rank P=0.017; Fig. 3B).
Additionally, at the protein level, high nuclear CIC expression was
also significantly associated with poorer OS (median OS, strong vs.
lower or absent, 25.4 vs. 79.6 months; log-rank P=0.012; Fig. 3C). Therefore, the data collectively
indicated that higher CIC expression is an unfavorable indicator
for OS time among patients with oligodendroglial tumors with
IDH mutations and no 1p/19q co-deletion.
Bioinformatics analysis
To explore the biological mechanisms for the
observed distinct prognostic role of CIC expression level,
the expression levels of previously identified genes targeted by
CIC [ETS variant (ETV)1, ETV4,
ETV5 and Cyclin D1] were compared within TCGA samples. It
was demonstrated that among IDH-mutant, 1p/19q co-deleted
oligodendroglial tumors, all the targets were significantly
upregulated in the lower expression group compared with the higher
expression groups (Fig. 4A),
suggesting a decrease in the transcriptional repressor activity of
CIC; whereas among the IDH-mutant, 1p/19q intact tumors, the
expression of the target genes was not significantly altered
(Fig. 4B), suggesting that the role
of CIC in this subset of gliomas may be independent of its
transcriptional regulatory activity. Furthermore, GSEA revealed
that among the IDH-mutant, 1p/19q co-deleted tumors, a reduction in
CIC expression was associated with the enrichment of gene
signatures involved in the biosynthesis of RNA and protein,
including translational initiation (P<0.0001) and biosynthetic
process (P<0.0001). By contrast, among those tumors without
1p/19q co-deletion, the increase in CIC expression was associated
with the enrichment of gene signatures involved in cell
proliferation, including regulation of mitosis (Normalized
enrichment score [NES]=−1.74, P=0.041), treatment resistance,
including nucleotide excision repair (NES=−1.52, P=0.050) and
epigenetic modification, including chromatin modification
(NES=−1.77, P=0.016).
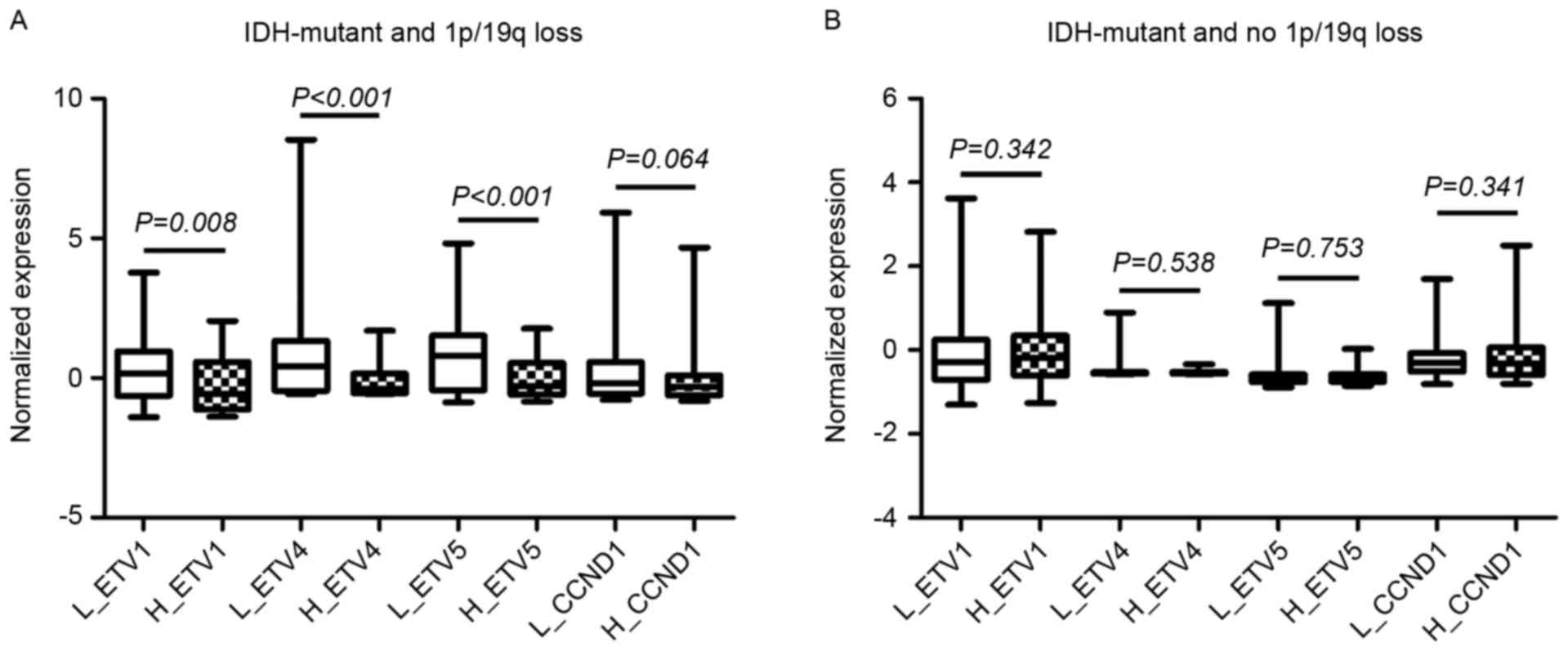 | Figure 4.The mRNA expression levels of CIC
target genes (ETV1, ETV4, ETV5 and
CCND1) within low and high CIC expression subgroups among
(A) IDH-mutant, 1p/19q co-deleted oligodendroglial tumors and (B)
IDH-mutant, 1p/19q not co-deleted oligodendroglial tumors in The
Cancer Genome Atlas dataset. CIC, capicua transcriptional
repressor; ETV, ETS variant; CCND1, Cyclin D1; L_, low CIC
expression group; H_, high CIC expression group; IDH, isocitrate
dehydrogenase. |
Alerted CIC expression is an
independent predictor for OS time among IDH-mutant, 1p/19q-intact
oligodendroglial tumors, but not among 1p/19q co-deleted
tumors
Within IDH-mutant and 1p/19q co-deleted
oligodendroglial tumors, a univariate Cox regression model revealed
that patient age and CIC expression groups (for the GEO and IHC
cohorts) were significantly associated with OS, whereas WHO grade,
histological type, Karnofsky performance status and CIC
mutations were not (Table II). The
largely consistent results from all cohorts supported the
robustness of this finding. However, the multivariate Cox model
demonstrated that only patient age was consistently an independent
indicator (Table II).
 | Table II.Results from the univariate and
multivariate Cox regression analyses among isocitrate
dehydrogenase-mutant, 1p/19q co-deleted oligodendroglial
tumors. |
Table II.
Results from the univariate and
multivariate Cox regression analyses among isocitrate
dehydrogenase-mutant, 1p/19q co-deleted oligodendroglial
tumors.
|
| TCGA | GEO | Xijing |
|---|
|
|
|
|
|
|---|
|
| Univariate Cox
model | Multivariate Cox
model | Univariate Cox
model | Multivariate Cox
model | Univariate Cox
model | Multivariate Cox
model |
|---|
|
|
|
|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| WHO grade | 4.46 | 0.003 | 3.61 | 0.012 | 0.89 | 0.841 |
|
|
1.16 | 0.853 |
|
|
|
| (1.67–11.87) |
| (1.31–9.88) |
| (0.27–2.77) |
|
|
| (0.24–5.52) |
|
|
|
| Histopathological
type | 0.34 | 0.296 |
|
| 0.78 | 0.808 |
|
| 27.20 | 0.366 |
|
|
|
| (0.05–2.57) |
|
|
| (0.10–6.07) |
|
|
|
(0.02–35,261.70) |
|
|
|
| Age | 3.17 | 0.006 | 2.87 | 0.033 | 5.78 | 0.008 | 4.63 | 0.036 | 14.22 | 0.013 | 13.73 | 0.018 |
|
| (1.46–9.72) |
| (1.09–7.60) |
| (1.59–20.94) |
| (1.10–19.44) |
| (1.74–116.48) |
| (1.57–120.02) |
|
| KPS | 0.90 | 0.103 |
|
| 0.99 | 0.470 |
|
|
0.97 | 0.126 |
|
|
|
| (0.80–1.02) |
|
|
| (0.96–1.02) |
|
|
| (0.94–1.01) |
|
|
|
| CIC
mutation | 0.78 | 0.586 |
|
| – | – |
|
| – | – |
|
|
|
| (0.32–1.92) |
|
|
|
|
|
|
|
|
|
|
|
| CIC expression
group | 0.03 | 0.194 |
|
| 0.27 | 0.018 | 0.63 | 0.444 |
0.24 | 0.040 | 0.26 | 0.074 |
|
| (0.00–5.80) |
|
|
| (0.09–0.80) |
| (0.19–2.08) |
| (0.06–0.94) |
| (0.06–1.14) |
|
The multivariate Cox model was not applied to
IDH-mutant, 1p/19q-intact oligodendroglial tumors as only
CIC expression group consistently reached statistical
significance in each cohort in the univariate Cox regression models
(Table III). The data suggested
that CIC expression groups may be an independent prognostic
factor in this subset of gliomas; however, further validation is
required.
 | Table III.Results from the univariate Cox
regression analysis among isocitrate dehydrogenase-mutant, 1p/19q
intact oligodendroglial tumors. |
Table III.
Results from the univariate Cox
regression analysis among isocitrate dehydrogenase-mutant, 1p/19q
intact oligodendroglial tumors.
|
| TCGA | GEO | Xijing |
|---|
|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| WHO grade | 1.00
(0.39–2.54) | 0.994 | 0.92
(0.12–7.35) | 0.940 | 2.81
(0.75–10.49) | 0.124 |
| Histopathologic
type | 0.97
(0.43–2.21) | 0.950 | 1.31
(0.41–4.18) | 0.644 | 0.54
(0.15–2.02) | 0.360 |
| Age | 0.91
(0.27–3.90) | 0.875 | 1.99
(0.51–7.76) | 0.323 | 0.82
(0.10–6.73) | 0.857 |
| KPS | 0.66
(0.13–3.29) | 0.610 | 0.97
(0.93–1.01) | 0.092 | 0.99
(0.94–1.04) | 0.662 |
| CIC
mutation | 0.45
(0.10–2.00) | 0.292 | – | – | – | – |
| CIC expression
group | 2.94
(1.29–6.67) | 0.010 | 4.15
(1.17–14.73) | 0.028 | 4.66
(1.24–17.48) | 0.023 |
Development of a simple classification
scheme based on CIC expression level for IDH-mutant
oligodendroglial tumors
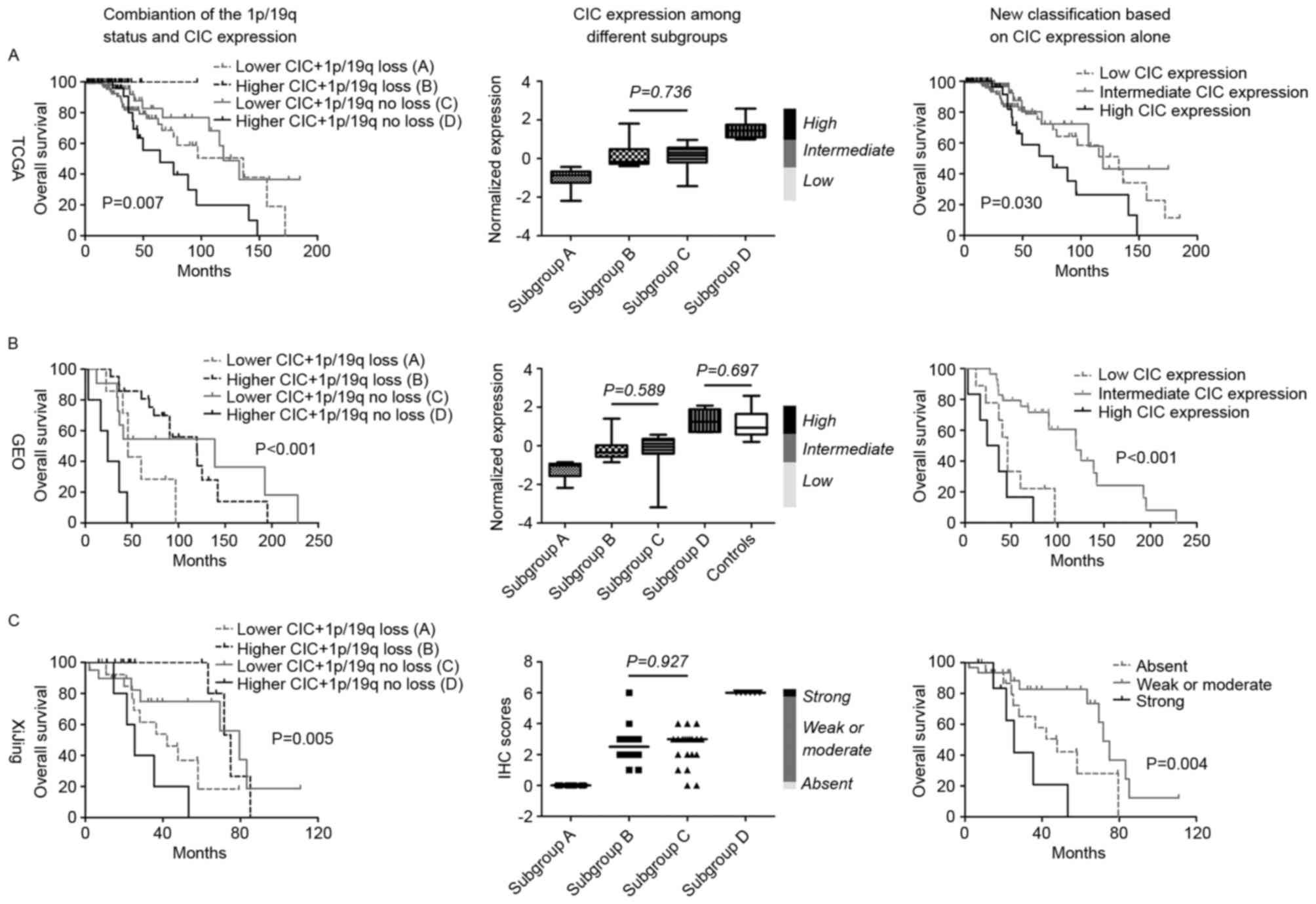
The combination of 1p/19q status and CIC
expression stratified all patients with IDH-mutant
oligodendroglial tumors into 4 prognostic subgroups: Subgroup
A (lower CIC expression in 1p/19q co-deleted tumors),
subgroup B (higher CIC expression in 1p/19q co-deleted
tumors), subgroup C (lower CIC expression in 1p/19q-intact
tumors) and subgroup D (higher CIC expression in
1p/19q-intact tumors). Survival analysis revealed that patients
within subgroups B and C exhibited the longest
overall survival times, and patients in subgroups D the
shortest, followed by subgroup A (Fig. 5). Accordingly, by comparing CIC
expression levels within the subgroups, subgroups A and
D demonstrated the lowest and highest CIC expression,
respectively, whereas subgroups B and C were observed
to possess similar, intermediate expression levels (Fig. 5). Therefore, a simple classification
scheme was devised based on the CIC expression of IDH-mutant
oligodendroglial tumors, with the thresholds of the upper
expression boundary of subgroup A and the lower expression
boundary of subgroup D. According to this novel
classification scheme, all patients were classified into 3 distinct
prognostic subgroups regardless of the 1p/19q status; patients with
intermediate CIC expression exhibited the longest survival times,
followed by those with low expression, whereas patients with high
CIC expression had the shortest survival times (Fig. 5). In summary, this novel prognostic
scheme may be useful for optimizing the risk classification of
IDH-mutant oligodendroglial tumors.
Discussion
The CIC gene has long been implicated in the
pathogenesis of oligodendroglial tumors (10). Recurrent mutations in this gene were
frequently observed in a subset of gliomas with IDH
mutations and 1p/19q co-deletion. Studies have demonstrated the
critical role of the genetic alterations to this gene in
determining tumor biological and clinical behaviors (3,4). However,
no consistent results have been reported regarding the association
between the genetic alterations and prognosis, which critically
compromises its clinical utility (7,9). The
results of the present study revealed that, despite the overall
reduced expression level, CIC mRNA expression was variable
across IDH-mutant oligodendroglial tumors. It was observed
that the CIC expression level was associated with different
tumor types (OA or OD), the 1p/19q status and patient ages. The
variable expression level also reflected the genetic status of
CIC; due to the frequent loss of one copy of chromosome arm
19q, subsequent inactivating mutations in gene coding regions of
the remaining copy may result in gene repression (2). Therefore, the CIC expression pattern may
be useful as an alternative to detecting somatic mutations in
predicting the prognosis of patients with oligodendroglial
tumors.
To the best of our knowledge, the present study
demonstrated for the first time the distinct prognostic value of
altered CIC expression depending on 1p/19q status among
IDH-mutant oliogendroglial tumors. Among tumors carrying the
1p/19q co-deletion, higher CIC expression (at mRNA and protein
levels) was a favorable indicator of survival, suggesting a
tumor-suppressive role in that subset of gliomas. The decreased
expression of CIC predominantly results from inactivating mutations
in gene coding regions and the rapid degradation of mutant proteins
(3). Decreased CIC expression is
associated with a decrease in CIC transcriptional repressor
activity and thus may have contributed to tumor aggression via the
induction of a transcriptional profile of high activity in
transcription and translation, in line with the GSEA results of the
present study. Recently, Gleize et al (4) demonstrated similar results regarding the
tumor-suppressive role of wild-type CIC, but with a focus on
its mutation status in IDH-mutant, 1p/19q co-deleted
oligodendroglial tumors. Mutant proteins were quickly adequately
degraded in those tumors, resulting in increased CIC expression in
certain cases (4). A previous study
identified that mutant proteins decrease cell clonogenicity and
slow tumor growth in the presence of mutant IDH proteins via
the regulation of cytosolic citrate metabolism (19). In such cases, tumors with the
increased expression of mutant CIC may be associated with favorable
OS. These observations support the hypothesis that quantifying CIC
expression may be more useful than determining its mutation status
for providing accurate prognostic information in this subset of
gliomas.
Until recently, only a small number of studies
focused on the roles of CIC alterations, including somatic
mutations and abnormal expression, in IDH-mutant,
1p/19q-intact oligodendroglial tumors. In line with previous
studies (3,8,9), the
results of the present study revealed a low incidence of CIC
mutations and no significant association between CIC
mutation and overall survival time in this subset of tumors, from
TCGA. In the present study, higher CIC expression (at mRNA
and protein levels) was associated with poor prognosis in
IDH-mutant, 1p/19q-intact oligodendroglial tumors. This suggested a
pro-oncogenic role for CIC, particularly the wild type, in
the molecular context of IDH mutations and 1p/19-intact.
Additionally, in this subset of gliomas, the tumor-promoting role
of CIC was possibly independent of its transcriptional
repression activity, as evidenced by the similar expression levels
of its targets among tumors with different levels of CIC
expression. GSEA analysis revealed that, in the presence of intact
1p/19q, increased CIC expression was associated with the
activation of proliferative pathways. Limited data is available
concerning the oncogenic role of wild-type CIC in cancer
biology. CIC was rarely observed to be mutated although it
was highly expressed in medulloblastoma, suggesting its relevance
in tumor genesis and progression (3,20).
Furthermore, a previous study demonstrated that wild-type CIC
protein may rescue the reduction in cell proliferation caused by
IDH1 mutations in the presence of intact 1p/19q (19). The aforementioned data, along with the
data of the present study, suggest a potential oncogenic role for
wild-type CIC in human cancer; however, further
investigation is required.
The equivocal histopathologic definition of
oligodendroglial tumors results in inefficiency in their management
(2). Molecular markers have
demonstrated promising value for optimizing the prediction of
prognosis for patients with these types of glioma (1). In the present study, the distinct
prognostic values of CIC expression with regard to the
1p/19q status among IDH-mutant oligodendroglial tumors was
demonstrated. The combination of altered CIC expression and the
1p/19q status yielded 4 distinct prognostic subgroups and provided
the optimized prediction of prognosis for patients with
IDH-mutant oligodendroglial tumors. Furthermore, by
comparing the expression levels of CIC within the subgroups,
it was identified that regardless of the 1p/19q status, CIC
expression was associated with distinct prognoses; tumors with
intermediate CIC expression were associated with the best outcomes,
whereas those with the lowest CIC expression had less favorable
outcomes and those with the highest expression had the poorest. In
addition, the results from the IHC cohort suggested that this novel
scheme may be accomplished by a simple but widely-used test method
in the clinical setting. Therefore, a novel classification scheme
based only on CIC expression level may exhibit clinical utility by
providing a simple way to rapidly estimate the prognosis of
patients with IDH-mutant oligodendroglial tumors without the
assessment of 1p/19q status.
The reproducibility and consistency of the distinct
prognostic value of CIC expression at the mRNA and protein
levels across the cohorts suggest that the results of the present
study are unlikely to be spurious due to technical artifacts or
sample bias. However, the significance of the present study has a
number of limitations. Neither mRNA quantification nor the IHC
method were able to distinguish between the mutant and wide-type
CIC, which brought uncertainty into data interpretation. In
addition, the sample size of the IHC cohort was limited, and
cytoplasmic CIC staining was not considered in IHC scoring.
In conclusion, to the best of our knowledge, the
present study demonstrated for the first time the distinct
prognostic value of CIC expression with regard to the 1p/19q
status among IDH-mutant oligodendroglial tumors. The novel
classification scheme based on the CIC expression level may be
clinically useful by providing a simple and rapid estimation for
the prognosis of patients with IDH-mutant oligodendroglial tumors.
This study also highlighted the necessity of further investigation
into the heterogeneous role of CIC, either in mutant or
wild-type forms, in oligodendroglial tumors.
Acknowledgements
The authors wish to thank Mrs. Jun-li Huo, and Mrs.
Juan Li (Department of Neurosurgery, Xijing Hospital) for
collecting and registering the samples and Dr Bo-lin Liu (Division
of Neurosurgery, Arrowhead Regional Medical Center, Colton, CA,
USA) for revising the manuscript. The present study was partially
funded by the Natural Science Basic Research Plan in Shaanxi
Province of China (grant no. 2014JM4099).
References
|
1
|
Cancer Genome Atlas Research Network, ;
Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA,
Rheinbay E, Miller CR, Vitucci M, et al: Comprehensive, integrative
genomic analysis of diffuse lower-grade gliomas. N Engl J Med.
372:2481–2498. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wesseling P, van den Bent M and Perry A:
Oligodendroglioma: Pathology, molecular mechanisms and markers.
Acta Neuropathol. 129:809–827. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sahm F, Koelsche C, Meyer J, Pusch S,
Lindenberg K, Mueller W, Herold-Mende C, von Deimling A and
Hartmann C: CIC and FUBP1 mutations in oligodendrogliomas,
oligoastrocytomas and astrocytomas. Acta Neuropathol. 123:853–860.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gleize V, Alentorn A, Connen de Kérillis
L, Labussière M, Nadaradjane AA, Mundwiller E, Ottolenghi C,
Mangesius S, Rahimian A, Ducray F, et al: CIC inactivating
mutations identify aggressive subset of 1p19q codeleted gliomas.
Ann Neurol. 78:355–374. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van Den Bent MJ, Brandes AA, Taphoorn MJ,
Kros JM, Kouwenhoven MC, Delattre JY, Bernsen HJ, Frenay M, Tijssen
CC, Grisold W, et al: Adjuvant procarbazine, lomustine and
vincristine chemotherapy in newly diagnosed anaplastic
oligodendroglioma: Long-term follow-up of EORTC brain tumor group
study 26951. J Clin Oncol. 31:344–350. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reuss DE, Sahm F, Schrimpf D, Wiestler B,
Capper D, Koelsche C, Schweizer L, Korshunov A, Jones DT, Hovestadt
V, et al: ATRX and IDH1-R132H immunohistochemistry with subsequent
copy number analysis and IDH sequencing as a basis for an
‘integrated’ diagnostic approach for adult astrocytoma,
oligodendroglioma and glioblastoma. Acta Neuropathol. 129:133–146.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiao Y, Killela PJ, Reitman ZJ, Rasheed
AB, Heaphy CM, de Wilde RF, Rodriguez FJ, Rosemberg S, Oba-Shinjo
SM, Nagahashi Marie SK, et al: Frequent ATRX, CIC, FUBP1 and IDH1
mutations refine the classification of malignant gliomas.
Oncotarget. 3:709–722. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yip S, Butterfield YS, Morozova O,
Chittaranjan S, Blough MD, An J, Birol I, Chesnelong C, Chiu R,
Chuah E, et al: Concurrent CIC mutations, IDH mutations and 1p/19q
loss distinguish oligodendrogliomas from other cancers. J Pathol.
226:7–16. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chan AK, Pang JC, Chung NY, Li KK, Poon
WS, Chan DT, Shi Z, Chen L, Zhou L and Ng HK: Loss of CIC and FUBP1
expressions are potential markers of shorter time to recurrence in
oligodendroglial tumors. Mod Pathol. 27:332–342. 2014.PubMed/NCBI
|
|
10
|
Bettegowda C, Agrawal N, Jiao Y, Sausen M,
Wood LD, Hruban RH, Rodriguez FJ, Cahill DP, McLendon R, Riggins G,
et al: Mutations in CIC and FUBP1 contribute to human
oligodendroglioma. Science. 333:1453–1455. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jimenez G, Shvartsman SY and Paroush Z:
The Capicua repressor-a general sensor of RTK signaling in
development and disease. J Cell Sci. 125:1383–1391. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fuller GN and Scheithauer BW: The 2007
revised World Health Organization (WHO) classification of tumours
of the central nervous system: Newly codified entities. Brain
Pathol. 17:304–307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen X, Yun J, Fei F, Yi J, Tian R, Li S
and Gan X: Prognostic value of nuclear hepatoma-derived growth
factor (HDGF) localization in patients with breast cancer. Pathol
Res Pract. 208:437–443. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gravendeel LA, Kouwenhoven MC, Gevaert O,
de Rooi JJ, Stubbs AP, Duijm JE, Daemen A, Bleeker FE, Bralten LB,
Kloosterhof NK, et al: Intrinsic gene expression profiles of
gliomas are a better predictor of survival than histology. Cancer
Res. 69:9065–9072. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Erdem-Eraslan L, Gravendeel LA, de Rooi J,
Eilers PH, Idbaih A, Spliet WG, den Dunnen WF, Teepen JL, Wesseling
P, Sillevis Smitt PA, et al: Intrinsic molecular subtypes of glioma
are prognostic and predict benefit from adjuvant procarbazine,
lomustine and vincristine chemotherapy in combination with other
prognostic factors in anaplastic oligodendroglial brain tumors: A
report from EORTC study 26951. J Clin Oncol. 31:328–336. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Woo HG, Park ES, Cheon JH, Kim JH, Lee JS,
Park BJ, Kim W, Park SC, Chung YJ, Kim BG, et al: Gene
expression-based recurrence prediction of hepatitis B virus-related
human hepatocellular carcinoma. Clin Cancer Res. 14:2056–2064.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:pp. 15545–15550. 2005;
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hothorn T and Zeileis A: Generalized
maximally selected statistics. Biometrics. 64:1263–1269. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chittaranjan S, Chan S, Yang C, Yang KC,
Chen V, Moradian A, Firme M, Song J, Go NE, Blough MD, et al:
Mutations in CIC and IDH1 cooperatively regulate 2-hydroxyglutarate
levels and cell clonogenicity. Oncotarget. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee CJ, Chan WI and Scotting PJ: CIC, a
gene involved in cerebellar development and ErbB signaling, is
significantly expressed in medulloblastomas. J Neurooncol.
73:101–108. 2005. View Article : Google Scholar : PubMed/NCBI
|