Introduction
Lung cancer is the most common cause of
cancer-associated mortality worldwide, accounting for 1.8 million
newly diagnosed cases and 1.6 million incidences of
cancer-associated mortality in 2012 (1). In recent years, epidermal growth factor
receptor tyrosine kinase inhibitors (EGFR-TKIs), including
gefitinib, erlotinib or afatinib, have been widely used in the
treatment of non-small cell lung cancer (NSCLC) patients, resulting
in markedly prolonged progression-free and overall survival times
compared with those of patients undergoing standard chemotherapies
(2–7).
However, the therapeutic effect of EGFR-TKIs is associated with the
mutation spectrum and status of EGFR (8). Deletions in exon 19 and L858R in exon 21
are known to sensitize patients to EGFR-TKI therapy (9,10), with
these mutations covering ~90% of oncogenic EGFR mutations
(11). By contrast, the T790M
substitution in exon 20 is the most common secondary mutation among
patients who acquire resistance to EGFR-TKIs (12). Osimertinib, a third-generation TKI
that specifically targets the T790M mutation, was introduced to in
2015 and has exhibited clinical efficacy in NSCLC patients with the
T790M mutation (13). Thus, in order
to select NSCLC patients suitable for EGFR-TKI therapy, it is
necessary to perform EGFR molecular testing with cancer
specimens from these patients (14).
In clinical practice, EGFR mutation status is
commonly analyzed by polymerase chain reaction (PCR)-based assays
using biopsy specimens from advanced NSCLC patients (14); however, cytological specimens are
often the only specimens available. Although tissue specimens
obtained by transbronchial or transcutaneous biopsies are
preferable (14), it is often
difficult to obtain sufficient cancer tissue to perform
morphological and molecular analyses. Cytological specimens may,
however, be useful diagnostic tools for these analyses and the
procedures used to obtain these specimens are less invasive than
those used to obtain biopsy specimens (15).
Previous studies have documented the use of
cytological specimens, including pleural effusion (PLE), for
EGFR mutation testing (16–19).
However, there are limited studies examining the utility of
bronchial lavage fluid (BLF), which is obtained following
transbronchial lung biopsy and usually contains fewer cancer cells
than PLE (20–22). In addition, data on the reported
performance of companion diagnostics, including the therascreen
EGFR RGQ PCR kit (the therascreen EGFR assay), primarily
rely on formalin-fixed paraffin-embedded (FFPE) tissue blocks from
resected or biopsy specimens and as such, there are limited data on
fresh cytological specimens (23).
The purpose of the present study was to compare the
efficiency of EGFR mutation detection between fresh
cytological samples (cell pellets and cell-free supernatants) and
FFPE cell blocks prepared from 1% EGFR mutation-positive
lung cancer cell line mixtures using the therascreen EGFR
assay. Furthermore, the utility of fresh BLF specimens from
patients with NSCLC was also validated against matched FFPE tissue
specimens in EGFR mutation detection using the therascreen
EGFR assay.
Materials and methods
Lung cancer cell line samples
A total of three types of cytological samples were
prepared: Fresh cell pellets, fresh supernatants, and FFPE cell
blocks from three series of lung cancer cell line samples,
including 1% EGFR mutant cells [sample (S) 1, 2, and 3]. The
following human lung cancer cell lines were used: PC9 [EGFR
E746_A750del (c.2235_2249del)] and A549 (EGFR wild-type).
The PC9 cell line was obtained directly from the Riken BioResource
Centre (Tsukuba, Japan). The A549 cell line was obtained directly
from the Japanese Cancer Research Bank (Tokyo, Japan). PC9 and A549
were mixed at a ratio of 1:99, so that the percentage of
EGFR-mutant cells in the mixture was 1%. The cell line
mixture was divided into three samples, S1, S2, and S3, and each
sample was washed with saline, centrifuged at 500 × g for 5 min at
room temperature, then divided into cell pellet samples (S1p, S2p,
and S3p) and cell-free supernatant samples (S1s, S2s, and S3s).
From the cell pellets, FFPE cell block samples (S1b, S2b, and S3b)
were produced through fixation with 20% neutral-buffered formalin
for 24 h at room temperature using a sodium alginate cell block
method (Fig. 1).
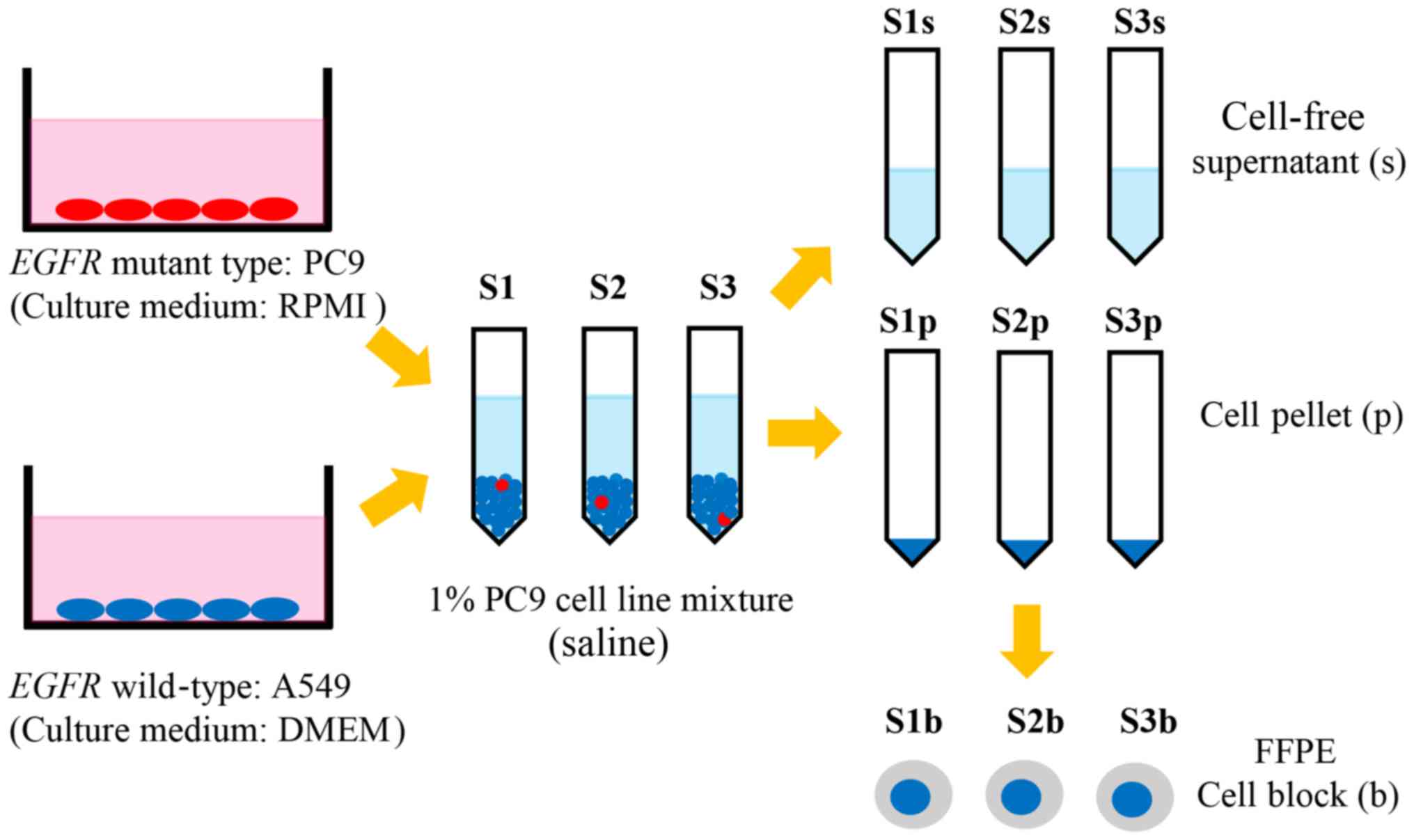 | Figure 1.Lung cancer PC9 [EGFR
E746_A750del (c.2235_2249del); red ovals] and A549 (EGFR
wild-type; blue ovals) cell lines were mixed at a ratio of 1:99,
meaning the percentage of EGFR mutant cells in the mixture
was 1%. The 1% EGFR mutation-positive mixture was divided
into samples S1, S2 and S3, which were washed with saline,
centrifuged and divided into S1p, S2p, and S3p, and S1s, S2s, and
S3s. From the cell pellets, S1b, S2b, and S3b were prepared.
EGFR, epithelial growth factor receptor; S1, sample 1; S1p,
S1 cell pellet; S1s, S1 cell-free supernatant; S1b, S1b
formalin-fixed paraffin embedded cell block. |
BLF and matched FFPE tissue specimens
from patients with NSCLC
Fresh BLF specimens were collected from 219 patients
who were clinically diagnosed with lung cancer between January 2014
and December 2014, and the specimens provided were used for
cytological diagnosis at the Department of Laboratory Medicine at
Shinshu University Hospital (Matsumoto, Japan). Following
cytological evaluation of specimens from 219 patients, specimens
that contained normal or benign cells (123 patients), malignant
lymphoma (1 patient), renal cell metastatic carcinoma (1 patient),
small cell carcinoma (8 patients), and squamous cell carcinoma (9
patients) were all excluded. The remaining 77 patients (age range,
41–85; median age, 69; 54 males and 23 females) whose specimens
were cytologically diagnosed as ‘primary lung adenocarcinoma’,
‘NSCLC-not otherwise specified (NSCLC-NOS)’, ‘suspicious for
malignancy’ (i.e., suspicious for adenocarcinoma or NSCLC-NOS), or
‘atypical cells’ meaning indefinite for neoplasia were enrolled in
this study. These patients are the same as those used in a previous
study by the same authors (22).
EGFR mutation status was analyzed using fresh
cell pellets from BLF specimens. Among the 77 patients, 49 patients
had EGFR mutation assay results from FFPE tissue specimens
obtained by surgical resection or biopsy. The assay results from
the BLF specimens were compared with those from the matching FFPE
tissue specimens. The present study was reviewed and approved by
the Medical Ethical Committee of the Shinshu University School of
Medicine. All patients provided written informed consent for
inclusion in the present study.
DNA extraction
DNA was extracted from fresh samples (cell pellets
and cell-free supernatants) using the QIAamp DNA Blood Mini kit
(Qiagen, Inc., Valencia, CA, USA) and the QIAamp DNA FFPE Tissue
kit (Qiagen, Inc.) for FFPE specimens, according to the
manufacturer's protocol. DNA concentration was quantified by
spectrophotometry using a NanoDrop ND100 instrument (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). For comparison of the three
types of cytological samples using cell lines, the therascreen
EGFR assays were performed following adjustment of the DNA
concentrations of cell pellets, cell-free supernatants, and FFPE
cell blocks to 4.5 ng/µl. For the validation study using BLF
specimens, the DNA concentrations were adjusted to <10
ng/µl.
EGFR mutation analysis
For EGFR mutation detection, the Rotor-Gene Q
5plex HRM instrument was used with the therascreen EGFR RGQ PCR kit
(therascreen EGFR assay; Qiagen, Inc.). This assay is
approved for use in the United States, Europe, Japan, and China,
and the kit is based on the amplification-refractory mutation
system (ARMS) and Scorpion PCR technology, which enable the
sensitive and selective site-specific detection of 29 types of
somatic mutations in EGFR (24). The reaction conditions were as
follows: 95°C for 15 min for 1 cycle; 95°C for 30 sec and 60°C for
60 sec for 40 cycles. The analysis was performed using the
Rotor-Gene Q series software, version 2.0.2 (Qiagen, Inc.). For
comparison between the three types of cytological samples (cell
pellets, cell-free supernatants and FFPE cell blocks) using cell
lines, the cycle quantification (Cq) value of the mutant allele in
each sample was compared using the therascreen EGFR
‘Deletions’ assay, which detects EGFR E746_A750del. For the
validation study comparing BLF specimens to FFPE tissue specimens,
the manufacturer-supplied cut-off delta Cq (ΔCq) values were used
to determine the result whether positive or negative for the
mutation in each EGFR mutation reaction.
Statistical analysis
For comparison of the cell line samples, the Quade
test (Statcel version 4; OMS Publishing, Tokorozawa, Japan) was
used to compare the differences in Cq values between the cell
pellets, cell-free supernatants, and FFPE cell blocks. The Quade
test indicates if there is a significant difference in the Cq
values among the three sample types: Cell pellets, cell-free
supernatants and FFPE cell blocks. The Cq values in three series
(S1, S2 and S3) were compared. P<0.05 was considered to indicate
a statistically significant difference.
Results
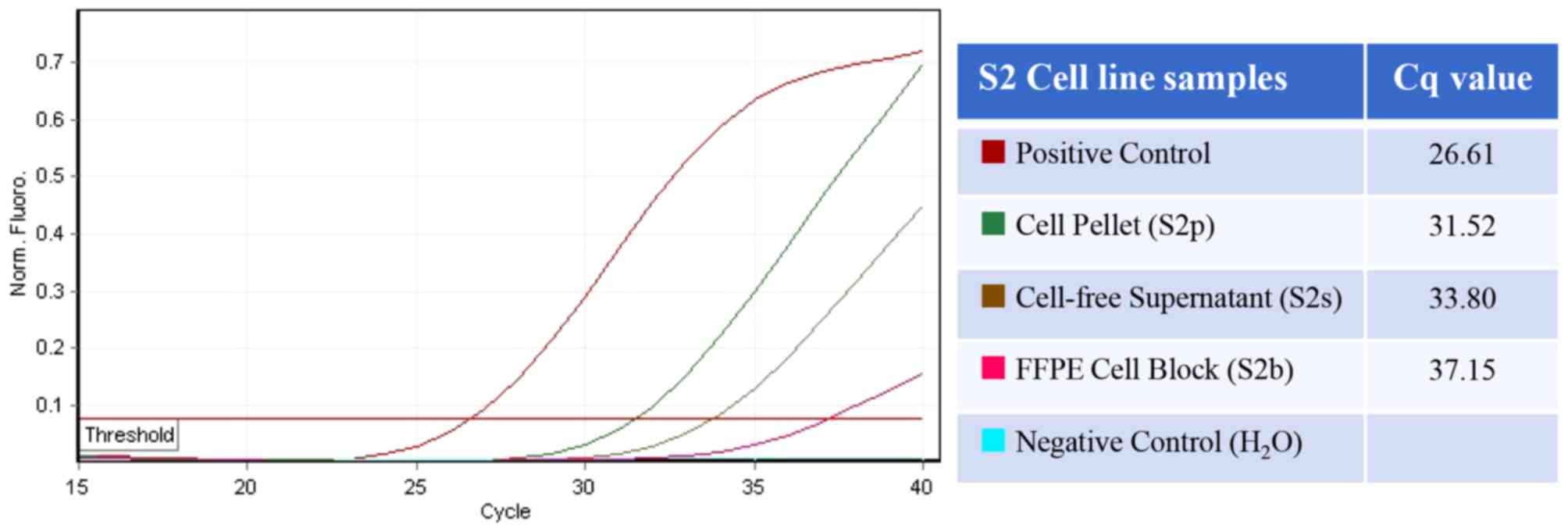
Comparison of Cq values between
samples
The Cq values of the cytological samples in the
three series, as determined by the therascreen EGFR assay,
are presented in Table I. For all
cell line samples excluding the FFPE cell block of S3, the
EGFR mutation (E746_A750del) was detected by therascreen
EGFR assay within 40 cycles. The average Cq values for
mutation detection for the three sample types in the three series
(S1, S2, and S3) were as follows: 29.58 for fresh cell pellets,
34.15 for fresh cell-free supernatants and 37.12 for FFPE cell
blocks. The Quade test revealed that the Cq values in each series
were significantly lower in cell pellets compared with that in the
other sample types, followed by cell-free supernatants and FFPE
cellblocks (P<0.05). The representative amplification curve and
Cq value for each sample in the S2 series are presented in Fig. 2.
 | Table I.Cq values of cell line samples in
therascreen EGFR assay. |
Table I.
Cq values of cell line samples in
therascreen EGFR assay.
| Cq value | Cell pellets | Cell-free
supernatants | FFPE cell
blocks | P-value |
|---|
| S1 | 24.93 | 34.43 | 35.17 | <0.05 |
| S2 | 31.52 | 33.80 | 37.15 |
|
| S3 | 32.28 | 34.21 | 39.05 |
|
| Average | 29.58 | 34.15 | 37.12 |
|
Assay results of 77 fresh BLF
specimens
EGFR mutation status was successfully
analyzed using cell pellets from all 77 BLF specimens. The
therascreen EGFR assay detected the exon 21 L858R point
mutation in 14 patients (18.2%), exon 19 deletions in 10 patients
(12.3%), and an exon 20 insertion in 1 patient (1.3%).
For the 49 patients who had EGFR mutation
assay results for matching FFPE tissue specimens, all EGFR
assay results were completely concordant between BLF cell pellets
and FFPE tissue specimens. The assay results for the BLF cell
pellets and matching FFPE tissue specimens in these 49 patients are
listed in Table II.
 | Table II.EGFR assay results of BLF cell
pellets and matching FFPE tissues. |
Table II.
EGFR assay results of BLF cell
pellets and matching FFPE tissues.
| EGFR
mutation status | BLF cell pellets, n
(%) | Matching FFPE
tissues, n (%) |
|---|
| Positive | 17 (34.7%) | 17 (34.7%) |
| Exon 19
deletions | 5 | 5 |
| Exon 21 L858R | 11 | 11 |
| Exon 20
insertions | 1 | 1 |
| Negative | 32 (65.3%) | 32 (65.3%) |
| Total | 49 | 49 |
The association between cytological diagnosis and
EGFR mutation status of BLF specimens is demonstrated in
Table III. EGFR mutations
were detected in specimens that were cytologically diagnosed as
‘primary lung adenocarcinoma’ or ‘NSCLC-NOS’ from 7 patients,
‘suspicious for malignancy’ from 6 patients, and ‘atypical cells’
from 4 patients.
 | Table III.EGFR mutation status and
cytological diagnosis of 49 BLF specimens. |
Table III.
EGFR mutation status and
cytological diagnosis of 49 BLF specimens.
| Cytological
diagnosis |
EGFR-mutation positive |
EGFR-mutation negative | Total |
|---|
| ADC or
NSCLC-NOS | 7 | 27 | 34 |
| Suspicious for
malignancy | 6 | 3 | 9 |
| Atypical cells | 4 | 2 | 6 |
| Total | 17 | 32 | 49 |
Discussion
Although the use of FFPE cell block specimens should
be prioritized for EGFR mutation testing, various
cytological specimens, including liquid-based cytology, smear,
fresh cell pellet and supernatant specimens are also suitable
(14–16,18,25,26).
However, the guidelines of the College of American
Pathologists/International Association for the Study of Lung
Cancer/Association for Molecular Pathology, currently recommend the
creation of FFPE cell blocks from cytological specimens in order to
perform morphological and immunohistochemical examination for
pathological diagnosis, and to stock them for additional molecular
diagnostic studies (14). A novel
EGFR mutation assay (the EGFR d-PCR assay) using
fresh liquid cytological specimens (cell pellets) was previously
validated, revealing that these specimens may contribute to the
rapid and simple point-of-care test (22). In the present study, the detection
efficiency for fresh cytological samples (not only cell pellets but
also cell-free supernatants) was demonstrated, as was the relative
EGFR mutation detection accuracy for fresh cell pellets from
BLF specimens versus FFPE tissue specimens.
When comparing cell pellets, cell-free supernatants
and FFPE cell blocks from cancer cell lines in the present study,
cell pellets had significantly lower Cq values compared with FFPE
cell blocks. This is because FFPE samples suffer from DNA
fragmentation and the formation of cross-links during the fixation
process (27); therefore, fresh cell
pellets are considered to contain DNA that is more efficacious for
mutation detection compared with FFPE cell blocks. In addition, DNA
derived from slices of FFPE cell blocks usually contain DNA from
only a small number of cancer cells, whereas fresh cytological
specimens contain DNA from a number of cancer cells (28). Hence, FFPE cell block samples are
assumed to exhibit a higher risk of false-negative outcomes
compared with fresh cell pellets from cytological specimens
(29,30).
In the present study, cell-free supernatant samples,
which are considered to contain less DNA than fresh cell pellets,
exhibited significantly better amplification efficiency compared
with FFPE cell blocks. The result from the present study indicated
that the fixation process has a strong negative impact on the
detection efficiency of EGFR mutations and is indicative of
the use of fresh cell-free supernatants of cytological specimens
for EGFR mutation detection assays. Previous studies have
reported the feasibility of detecting EGFR mutations with
fresh cell-free supernatants of PLEs by direct sequencing (31), mutant-enriched PCR (19) and Scorpion ARMS (17). Zhang et al (19) revealed a good concordance between
EGFR mutation assay results from cell pellets and those from
matching cell-free fluids following mutant-enriched PCR. By using
highly sensitive methods for EGFR mutation detection,
performing EGFR assays with cell-free supernatants of BLF
specimens is possible and may be useful, as the cell pellets, which
enable morphological and multiple molecular analyses are
preserved.
In the validation of the EGFR mutational
status of BLF specimens using cell pellets from 77 patients with
NSCLC, all EGFR mutational statuses were successfully
analyzed and there were no specimens that failed to amplify. The
assay results for all 49 BLF cell pellets with matching FFPE tissue
specimens were completely concordant between the two specimen
types. Goto et al (16)
reported an excellent interassay concordance between 29
bronchofiberscopic brushing cytological specimens and matching FFPE
specimens using Scorpion ARMS, PCR-Invader, PNA-LNA PCR clamp, and
Cycleave PCR, with concordance rates of 93.1–96.6% (κ-coefficients,
0.86–0.93). Khode et al (32)
compared 37 paired cytological smears with matched FFPE surgical
tissue specimens from the same anatomical sites using
pyrosequencing and RT-PCR platforms, and additionally reported a
concordance rate of 97% between the two sample types. The present
study also demonstrated the accuracy of the EGFR assay using
fresh cytological specimens for EGFR mutation detection,
with high concordance following the therascreen EGFR assay
results of FFPE tissue specimens.
Among the 77 fresh BLF specimens, EGFR
mutations were even detected in specimens containing only a few
cancer cells, which were cytologically diagnosed as ‘atypical
cells’ (i.e., indefinite for neoplasia). As was demonstrated by
comparing different cytological samples from cell lines, use of the
EGFR mutation assay with fresh cell pellets resulted in
higher sensitivity of mutation detection compared with the use of
FFPE cell blocks, even with a small amount of mutant DNA derived
from a few cancer cells being detected by the assay. Furthermore,
the use of BLF specimens may also markedly reduce the time and
effort required for DNA extraction compared with the use of FFPE
specimens, as BLF specimens do not require deparaffinization or
reversal of formaldehyde-induced nucleic acid modification. Thus,
the EGFR mutation assay with fresh BLF specimens may
represent a screening method for diagnosing whether atypical cells
are cancerous or not, particularly when morphological diagnosis is
difficult owing to the presence of insufficient material.
Despite the fact that fresh BLF specimens contain
DNA that is more efficacious for an EGFR mutation detection
assay, they also usually contain more non-cancerous cells,
including inflammatory cells and benign epithelial cells, compared
with tissue specimens. The amount of DNA from non-cancerous cells
affects the quality of the mutation-specific PCR assay, which may
lead to false-negative results. Furthermore, for the current major
companion diagnostics for EGFR-TKIs, including the therascreen
EGFR assay, the cut-off values have been determined for DNA
samples extracted from FFPE tissue or cell block specimens, as
fresh cytological specimens are not generally used for testing.
Therefore, the results of testing fresh cytological specimens
should be interpreted carefully, and priority should be given to
FFPE tissue specimens or cell blocks if they contain sufficient
cancer cells.
To conclude, EGFR mutation detection assays
using fresh BLF specimens offer a sensitive, accurate, simple and
time-saving method for detection of EGFR mutations, even
when sufficient cancer tissues or cytological specimens are not
obtained. This method enables the full use of specimens from NSCLC
patients for multiple molecular analyses.
Acknowledgements
The present study was partly supported by the Japan
Society for the Promotion of Science Grants-in-Aid for Scientific
Research, commonly termed KAKENHI (grant no. 25460434).
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
EGFR
|
epidermal growth factor receptor
|
|
EGFR-TKIs
|
EGFR tyrosine kinase inhibitors
|
|
BLF
|
bronchial lavage fluid
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global Cancer Statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: Gefitinib or chemotherapy for non-small-cell lung cancer
with mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mitsudomi T, Morita S, Yatabe Y, Negoro S,
Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, et
al: Gefitinib versus cisplatin plus docetaxel in patients with
non-small-cell lung cancer harbouring mutations of the epidermal
growth factor receptor (WJTOG3405): An open label, randomised phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou C, Wu YL, Chen G, Feng J, Liu XQ,
Wang C, Zhang S, Wang J, Zhou S, Ren S, et al: Erlotinib versus
chemotherapy as first-line treatment for patients with advanced
EGFR mutation-positive non-small-cell lung cancer (OPTIMAL,
CTONG-0802): A multicentre, open-label, randomised, phase 3 study.
Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rosell R, Carcereny E, Gervais R,
Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R,
Pallares C, Sanchez JM, et al: Erlotinib versus standard
chemotherapy as first-line treatment for European patients with
advanced EGFR mutation-positive non-small-cell lung cancer
(EURTAC): A multicentre, open-label, randomised phase 3 trial.
Lancet Oncol. 13:239–246. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang
Y, Li W, Hou M, Shi JH, Lee KY, et al: Afatinib versus cisplatin
plus gemcitabine for first-line treatment of Asian patients with
advanced non-small-cell lung cancer harbouring EGFR mutations
(LUX-Lung 6): An open-label, randomised phase 3 trial. Lancet
Oncol. 15:213–222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang JC, Wu YL, Schuler M, Sebastian M,
Popat S, Yamamoto N, Zhou C, Hu CP, O'Byrne K, Feng J, et al:
Afatinib versus cisplatin-based chemotherapy for EGFR
mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6):
Analysis of overall survival data from two randomised, phase 3
trials. Lancet Oncol. 16:141–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fukuoka M, Wu YL, Thongprasert S,
Sunpaweravong P, Leong SS, Sriuranpong V, Chao TY, Nakagawa K, Chu
DT, Saijo N, et al: Biomarker analyses and final overall survival
results from a phase III, randomized, open-label, first-line study
of gefitinib versus carboplatin/paclitaxel in clinically selected
patients with advanced non-small-cell lung cancer in Asia (IPASS).
J Clin Oncol. 29:2866–2874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Naidoo J, Sima CS, Rodriguez K, Busby N,
Nafa K, Ladanyi M, Riely GJ, Kris MG, Arcila ME and Yu HA:
Epidermal growth factor receptor exon 20 insertions in advanced
lung adenocarcinomas: Clinical outcomes and response to erlotinib.
Cancer. 121:3212–3220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee VH, Tin VP, Choy TS, Lam KO, Choi CW,
Chung LP, Tsang JW, Ho PP, Leung DK, Ma ES, et al: Association of
Exon 19 and 21 EGFR mutation patterns with treatment outcome after
first-line tyrosine kinase inhibitor in metastatic non-small-cell
lung cancer. J Thorac Oncol. 8:1148–1155. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mitsudomi T, Kosaka T and Yatabe Y:
Biological and clinical implications of EGFR mutations in lung
cancer. Int J Clin Oncol. 11:190–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuiper JL, Heideman DA, Thunnissen E, Paul
MA, van Wijk AW, Postmus PE and Smit EF: Incidence of T790M
mutation in (sequential) rebiopsies in EGFR-mutated NSCLC-patients.
Lung Cancer. 85:19–24. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang S, Cang S and Liu D: Third-generation
inhibitors targeting EGFR T790M mutation in advanced non-small cell
lung cancer. J Hematol Oncol. 9:342016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lindeman NI, Cagle PT, Beasley MB, Chitale
DA, Dacic S, Giaccone G, Jenkins RB, Kwiatkowski DJ, Saldivar JS,
Squire J, et al: Molecular testing guideline for selection of lung
cancer patients for EGFR and ALK tyrosine kinase inhibitors
guideline from the College of American Pathologists, International
Association for the Study of Lung Cancer, and Association for
Molecular Pathology. J Mol Diagn. 15:415–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ellison G, Zhu GS, Moulis A, Dearden S,
Speake G and McCormack R: EGFR mutation testing in lung cancer: A
review of available methods and their use for analysis of tumour
tissue and cytology samples. J Clin Pathol. 66:79–89. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Goto K, Satouchi M, Ishii G, Nishio K,
Hagiwara K, Mitsudomi T, Whiteley J, Donald E, McCormack R and Todo
T: An evaluation study of EGFR mutation tests utilized for
non-small-cell lung cancer in the diagnostic setting. Ann Oncol.
23:2914–2919. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kimura H, Fujiwara Y, Sone T, Kunitoh H,
Tamura T, Kasahara K and Nishio K: High sensitivity detection of
epidermal growth factor receptor mutations in the pleural effusion
of non-small cell lung cancer patients. Cancer Sci. 97:642–648.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin J, Gu Y, Du R, Deng M, Lu Y and Ding
Y: Detection of EGFR mutation in supernatant, cell pellets of
pleural effusion and tumor tissues from non-small cell lung cancer
patients by high resolution melting analysis and sequencing. Int J
Clin Exp Pathol. 7:8813–8822. 2014.PubMed/NCBI
|
|
19
|
Zhang X, Zhao Y, Wang M, Yap WS and Chang
AY: Detection and comparison of epidermal growth factor receptor
mutations in cells and fluid of malignant pleural effusion in
non-small cell lung cancer. Lung Cancer. 60:175–182. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamaguchi F, Kugawa S, Tateno H, Kokubu F
and Fukuchi K: Analysis of EGFR, KRAS and P53 mutations in lung
cancer using cells in the curette lavage fluid obtained by
bronchoscopy. Lung Cancer. 78:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sakamoto T, Kodani M, Takata M, Chikumi H,
Nakamoto M, Nishii-Ito S, Ueda Y, Izumi H, Makino H, Touge H, et
al: A novel point-of-care system for high-speed real-time
polymerase chain reaction testing for epidermal growth factor
receptor mutations in bronchial lavage fluids after transbronchial
biopsy in patients with non-small cell lung cancer. Int J Oncol.
46:1473–1480. 2015.PubMed/NCBI
|
|
22
|
Asaka S, Yoshizawa A, Matsuda K, Yamaguchi
A, Yamamoto H, Shiina T, Nakata R, Ogawa K, Zhang M and Honda T: A
novel, rapid point-of-care test for lung cancer patients to detect
epidermal growth factor receptor gene mutations by using real-time
droplet-PCR and fresh liquid cytology specimens. Oncol Rep.
37:1020–1026. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Syed YY: Therascreen® EGFR RGQ
PCR Kit: A companion diagnostic for afatinib and gefitinib in
non-small cell lung cancer. Mol Diagn Ther. 20:191–198. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vallée A, Le Loupp AG and Denis MG:
Efficiency of the Therascreen® RGQ PCR kit for the
detection of EGFR mutations in non-small cell lung carcinomas. Clin
Chim Acta. 429:8–11. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lozano MD, Zulueta JJ, Echeveste JI,
Gúrpide A, Seijo LM, Martín-Algarra S, Del Barrio A, Pio R, Idoate
MA, Labiano T and Perez-Gracia JL: Assessment of epidermal growth
factor receptor and K-ras mutation status in cytological stained
smears of non-small cell lung cancer patients: Correlation with
clinical outcomes. Oncologist. 16:877–885. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Reynolds JP, Tubbs RR, Minca EC, MacNamara
S, Almeida FA, Ma PC, Pennell NA and Cicenia JC: EGFR mutational
genotyping of liquid based cytology samples obtained via fine
needle aspiration (FNA) at endobronchial ultrasound of non-small
cell lung cancer (NSCLC). Lung Cancer. 86:158–163. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dedhia P, Tarale S, Dhongde G, Khadapkar R
and Das B: Evaluation of DNA extraction methods and real time PCR
optimization on formalin-fixed paraffin-embedded tissues. Asian Pac
J Cancer Prev. 8:55–59. 2007.PubMed/NCBI
|
|
28
|
Gailey MP, Stence AA, Jensen CS and Ma DQ:
Multiplatform comparison of molecular oncology tests performed on
cytology specimens and formalin-fixed, paraffin-embedded tissue.
Cancer Cytopathol. 123:30–39. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Harada S, Agosto-Arroyo E, Levesque JA,
Alston E, Janowski KM, Coshatt GM and Eltoum IA: Poor cell block
adequacy rate for molecular testing improved with the addition of
Diff-Quik-stained smears: Need for better cell block processing.
Cancer Cytopathol. 123:480–487. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu H, Sun W, Zhang G and Cheng Y:
Detection of epidermal growth factor receptor mutation in
non-small-cell lung carcinoma using cytological and histological
specimens. J BUON. 20:142–145. 2015.PubMed/NCBI
|
|
31
|
Kimura H, Fujiwara Y, Sone T, Kunitoh H,
Tamura T, Kasahara K and Nishio K: EGFR mutation status in
tumour-derived DNA from pleural effusion fluid is a practical basis
for predicting the response to gefitinib. Br J Cancer.
95:1390–1395. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Khode R, Larsen DA, Culbreath BC, Parrish
S, Walker KL, Sayage-Rabie L, Beissner RS and Rao A: Comparative
study of epidermal growth factor receptor mutation analysis on
cytology smears and surgical pathology specimens from primary and
metastatic lung carcinomas. Cancer Cytopathol. 121:361–369. 2013.
View Article : Google Scholar : PubMed/NCBI
|