Introduction
Cancers are among the leading causes of mortality
worldwide, with 8.2 million cancer-related deaths in 2014. Among
different cancers, lung cancer has the highest mortality rate with
1.59 million deaths; this is more than twice the number of
hepatocellular carcinoma deaths, which is the second most fatal
form of cancer (1). Lung cancer
patients have poor prognosis and at initial hospital visit are
often diagnosed at an advanced stage, beyond the possibility of
surgical intervention (2).
Cytotoxic chemotherapeutic agents are usually
administered for advanced lung cancer therapy and are chosen
according to histological subtype. Recently, molecular targeted
agents and immune checkpoint inhibitors have been developed and
have been in clinical use since the 2000s (3). In non-small cell lung cancer, including
adenocarcinoma (ADC), which represents 40% of all lung cancers,
several types of driver gene mutation that promote oncogenic
transformation and tumor growth by aberrant activation of
proliferation signaling pathways have been identified (4). These driver genes are proposed as novel
candidates for molecular targeted therapy. Epidermal growth factor
receptor (EGFR) activating mutations lead to auto-phosphorylation
and promote EGFR/KRAS/MEK/ERK signaling (5). Aberrant anaplastic lymphoma kinase (ALK)
protein, produced by an ALK fusion gene, drives MEK/ERK and
PI3K/Akt pathways (6). EGFR-tyrosine
kinase inhibitor (TKI) and an ALK inhibitor against the above two
aberrant signals prevent tumor progression with a high response
rate of 56.0–70.0%, while cytotoxic chemotherapy or immune
checkpoint inhibition are successful only in 19.0–34.1% of cases
(7–11). Thus, these precision medicines based
on genetic or molecular features of lung ADC can provide therapy
with a high response rate and fewer adverse effects (7–11).
However, 29.2–40.0% of lung ADC patients have no targetable genetic
features (4,12,13). There
is an urgent need to discover novel biomarkers and therapeutic
targets, and studies are ongoing to establish targeted therapy for
rare driver gene mutations of malignancy (4).
We have identified and reported family with sequence
similarity 83, member B (FAM83B) as a novel diagnostic and
prognostic marker for lung squamous cell cancer (SqCC) (14). Comprehensive gene expression analysis
using cDNA microarray analysis and immunohistochemistry showed lung
SqCC expressed higher levels of FAM83B compared with lung
ADC or adjacent normal lung tissue, and correlated with patient
prognosis (14). FAM83B is
also overexpressed in several other types of cancer, such as
breast, ovary, bladder, and lung, and is associated with tumor
proliferation (15). Additionally,
induction of FAM83B in human mammary epithelial cells
resulted in neoplastic growth by increasing mitogen-activated
protein kinase (MAPK) signaling, while induction in human breast
cancer cell lines resulted in TKI resistance (15). Aberrant EGFR signals and downstream
signaling play an important role in targeted therapy for lung ADC;
therefore, we assumed that FAM83B also correlates with tumor
oncogenesis and growth in lung ADC. Here, we show an association
between FAM83B expression in lung ADC and demographics and
clinicopathological features.
Materials and methods
Ethics statement
This study was conducted with approval of the Ethics
Committee of Fukushima Medical University (approval no. 2775). The
participants' human rights and welfare were defended in accordance
with the Declaration of Helsinki. Written informed consent was
obtained from all participants.
Case selection
We identified 216 patients who underwent lung
resection at Fukushima Medical University between January 2008 and
June 2015 and who were pathologically diagnosed as primary lung
ADC. FAM83B mRNA levels were determined in matched lung
tumor and adjacent normal lung tissue and compared with clinical
features and prognosis. The data collected were; age at surgery,
sex, smoking history, presence of activating EGFR mutation
in the tumor, histological ADC subtype, tumor size, lymph node
metastasis, distant metastasis, pleural invasion, lymphovascular
invasion, vascular invasion, date of surgery, date of recurrence,
last confirmed survival date, and date of death. Disease-free
survival (DFS) was defined as the time from surgery to the first
recurrence or death. Overall survival (OS) was defined as the time
from surgery to death. To ensure a sufficient observation period,
prognostic analyses were performed for patients who underwent
complete resection up to December 2013, and who were followed up
for 5 years. Patients who had an additional advanced malignant
history within 5 years before lung resection, died of postoperative
complications, or who were followed up for less than 12 months were
excluded. In total, 126 patients were analyzed.
Comprehensive gene expression
analysis
Matched tumor and adjacent normal lung tissue
samples, 7 mm3 in size, were excised from surgical
specimens and frozen in liquid nitrogen. Frozen samples were
processed for total RNA extraction using ISOGEN (Nippon Gene,
Tokyo, Japan). As a control, common reference RNA was prepared by
mixing equal amounts of total RNA extracted from 22 human cell
lines to reduce cell type-specific expression bias (16). Synthetic 80mer polynucleotide probes
representing 14,400 human transcripts (MicroDiagnostic, Tokyo,
Japan) were arrayed using a custom arrayer. Labeled cDNA was
synthesized from 5 µg of sample RNA using SuperScript II (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) and Cyanine 5-dUTP
(PerkinElmer, Inc., Waltham, MA, USA), while Cyanine 3-dUTP
(PerkinElmer, Inc.)-labeled cDNA was synthesized from 5 µg of human
common reference RNA. Hybridization was performed using a Labeling
and Hybridization kit (Microdiagnostic). Signals were measured
using a GenePix 4000B Scanner (Axon Instruments; Molecular Devices,
LLC, Sunnyvale, CA, USA), and then processed into primary
expression ratios (ratio of Cyanine-5 intensity of each sample to
the Cyanine-3 intensity of human common reference RNA). Each ratio
was normalized by multiplication with normalization factors using
GenePix Pro 3.0 software (Axon Instruments; Molecular Devices,
LLC). The primary expression ratios were converted into
log2 fold changes (designated log ratios). An expression
ratio of 1 (i.e., log ratio of 0) was assigned to spots that
exhibited fluorescence intensities under detection limits, and we
included these in the calculation of signal averages. Data were
processed using Microsoft Excel software (Microsoft Corporation,
Redmond, WA, USA) and the MDI gene expression analysis software
package (MicroDiagnostic, Inc., Tokyo, Japan). mRNA expression data
related to FAM83B were extracted for this study.
Preparation of cell lines
HLC-1 cells were purchased from Riken Cell Bank
(Saitama, Japan). NCI-H2347, NCI-H1975 and MCF-7 cells were
purchased from American Type Culture Collection (Manassas, VA,
USA). HLC-1 cells were grown in Ham's F12 medium (087–08335; Wako
Pure Chemical Industries, Ltd., Osaka, Japan). NCI-H2347 and
NCI-H1975 cells were grown in RPMI-1640 medium (R8758;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), MCF-7 cells were
grown in DMEM medium (Wako Pure Chemical Industries, Ltd.). All
media contained 10% fetal bovine serum (Nichirei Biosciences,
Tokyo, Japan) and 1% penicillin-streptomycin (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany). Cells were cultured at 37°C in a
humidified atmosphere of 5% CO2.
siRNA preparation
siRNAs against FAM83B were purchased
(Hs_FAM83B_8 and Hs_FAM83B_9 FlexiTube siRNA; Qiagen GmbH,, Hilden,
Germany). Sequences of siRNAs were: Hs_FAM83B_8 (siRNA-8):
5′-CAGGAACGAGTTTCAGACTTT-3′, and Hs_FAM83B_9 (siRNA-9):
5′-TCCCGTTATTTGACAACTCAA-3′.
As a negative control, AllStars negative control
siRNA (Qiagen GmbH,) was used. As a positive control, AllStars Hs
Cell Death siRNA (Qiagen GmbH,) was used.
siRNA transfection efficiency
assay
For 96-well siRNA transfections, 0.3 µl of
Lipofectamine RNAiMAX (Thermo Fisher Scientific, Inc.) in 10 µl of
serum-free Opti-MEM (Thermo Fisher scientific, Inc.) was added to
preplated siRNAs in each well and incubated for 5 min at room
temperature. MCF7, HLC-1, H2347, or H1975 cells were added at
1.0×104 to each well. After incubation for 72 h, 10 µl
of Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan) was
added and absorbance at 450 nm measured 4 h later using a Multiskan
GO (Thermo Fisher scientific, Inc.). Each test was replicated three
times.
RNA interference and cell
proliferation assay
According to a transfection efficiency test, RNA
interference experiments (RNAi) were performed using siRNA-9.
siRNA-9 (final concentration of 2.5 nM) and 7.5-µl Lipofectamine
RNAiMAX were mixed in 100 µl of serum-free Opti-MEM in a
microcentrifuge tube, then added within 20 min to cells in 6-well
plates. RNAi was performed using 1×105 cells/well for
HLC-1, and 5×104 cells/well for H1975 and replicated
three times. To determine the silencing effects of siRNA against
FAM83B, cell numbers were counted after transfection using
Clone select imager (Molecular devices Japan, Tokyo, Japan).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cell lines using TRIzol
reagent (Thermo Fisher Scientific, Inc.) and a PureLink RNA Mini
Kit (Thermo Fisher Scientific, Inc.) according to the
manufacturers' instructions. RNA quantity was assessed using a
Nanodrop UV-Vis Spectrophotometer (Thermo Fisher Scientific, Inc.),
and samples with a 260/280 nm absorbance ratio of 1.8 or larger
were adopted as eligible for RT-PCR. Relative mRNA expression was
determined by RT-PCR. One-step RT-qPCR using a Taqman
RNA-to-CT 1-Step kit (Thermo Fisher Scientific, Inc.)
was performed according to the manufacturer's instructions. To
detect FAM83B mRNA, Taqman gene expression assays
(Hs00289694_m1; Thermo Fisher Scientific, Inc.) were used. As an
endogenous control, glyceraldehyde-3-phosphate dehydrogenase
(GAPDH, Hs02758991_g1; Thermo Fisher Scientific, Inc.) was
used. Forty cycles of amplification were performed for each
triplicated sample. The ΔΔCq method was applied for quantitative
evaluation (17). Cycle
quantification (Cq) values were calculated by Step One Plus
software version 2.3 (Thermo Fisher Scientific, Inc.). ΔCq was
defined as the difference between FAM83B Cq and GAPDH
Cq, and ΔΔCq was defined as the ratio to the endogenous control
sample. Signals undetected after 40 cycles were considered to have
an expression of zero.
SDS-page
Cell lysates were prepared by homogenization of
cells in RIPA lysis buffer (SC-24948; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA), using a Polytron homogenizer (Sonifier
SFX250; Emerson Electric Co., St. Louis, MO, USA) at 4°C. After
centrifugation at 10,000 × g for five min at 4°C, supernatants were
mixed with an equal volume of Sample buffer (2X Laemmli Sample
Buffer; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
2-mercaptoethanol (Bio-Rad; 200:1) was then added and samples
heated for three min at 100°C. Five micrograms of each sample were
then loaded on a polyacrylamide gel (Supersep ace 5–20%; Wako Pure
Chemical Industries, Ltd.) and electrophoresis was performed to
separate proteins (18).
Western blotting
Proteins were transferred to a polyvinylidene
difluoride membrane (Immobilon; Merck KGaA) in Towbin transfer
buffer (25 mM Tris base, 192 mM glycine, 0.1% SDS, 20% methanol)
(19). The membranes were then
blocked with 5% skimmed milk in PBS (0.137 M NaCl, 2.6 mM KCl, 1.8
mM KH2PO4, 8.1 mM
Na2HPO4/12H2O) and incubated
overnight in primary antibody solution at 4°C. Anti-FAM83B
antibodies (1:1,000; PA5-28548; ThermoFisher scientific, or
1:2,000; HPA031464; Atlas Antibodies AB, Stockholm, Sweden) or an
anti-GAPDH antibody (1:2,500; no. 2118; Cell Signaling Technology,
Inc., Danvers, MA, USA) were used as primary antibodies. Membranes
were then incubated with secondary antibody (anti-rabbit IgG,
Horseradish Peroxidase-Linked species-specific whole antibody
(1:20,000; GE Healthcare UK Ltd., Amersham, UK). The
chemiluminescent signals were captured with the ImageQuant LAS 4000
system (GE Healthcare UK Ltd.) using ECL select Western Blotting
Detection Reagent (GE Healthcare UK Ltd.) according to the
manufacturer's instructions.
Statistical analysis
Statistical analyses were performed using SPSS 21.0
(IBM Corp., Armonk, NY, USA). The patient cohort was divided into
two subgroups according to high or low FAM83B expression
with the log ratio of zero as the boundary. Patients were divided
into two groups according to median age. Tumor (T), Nodes (N), and
Metastasis (M) (TNM) factors of lung cancer were classified
according to the Union for International Cancer Control 7th edition
(20). T factor was not adopted but
tumor size and pleural invasion were. Continuous variables were
compared by two-tailed t-tests or one-way ANOVA, and
categorical variables were compared by the Chi-square test or
Fisher's exact test, as appropriate. Multivariate analyses using a
binary logistic regression model were performed to evaluate
independent predictors of FAM83B expression. DFS and OS were
estimated using the Kaplan-Meier method, and survival curves were
compared using log-rank tests. Variables that were suitable for a
Cox proportional hazards univariate model with significance were
analyzed by a multivariate model to adjust for potential
confounders. P<0.05 was considered to indicate a statistically
significant difference.
Results
FAM83B is highly expressed in ADC with
wild type-EGFR
This study included 119 male and 97 female patients,
with a mean age of 68.5 years (range 26–87 years). Up to 111
(51.4%) were current or former smokers, and 118 (54.6%) had
wild-type EGFR ADC. FAM83B tended to be expressed at
higher levels in solid subtypes while lower FAM83B
expression was observed in lepidic pattern tumors that were less
aggressive. Mean tumor size was 2.9 cm (range 0.8–14.0). The
clinicopathological characteristics of patients according to
FAM83B expression in tumor tissue are summarized in Table I. Univariate analysis showed that
higher FAM83B expression correlates with males (P=0.007),
smoking history (P=0.007), and wild-type EGFR tumors
(P<0.001). Multivariate analysis showed wild-type EGFR
tumors correlate with FAM83B expression (P<0.001)
(Table II).
 | Table I.Clinicopathological and genetic
features of lung adenocarcinoma patients according to fam83b
expression in tumor tissue. |
Table I.
Clinicopathological and genetic
features of lung adenocarcinoma patients according to fam83b
expression in tumor tissue.
|
| FAM83B
expression |
|
|---|
|
|
|
|
|---|
| Variable | High (n=55)
(%) | Low (n=161)
(%) | P-value |
|---|
| Age |
|
| 0.271 |
| ≤69
years old | 25 (45.5) | 87 (54.0) |
|
| ≥70
years old | 30 (54.5) | 74 (46.0) |
|
| Sex |
|
| 0.006 |
|
Male | 39 (70.9) | 80 (49.7) |
|
|
Female | 16 (29.1) | 81 (50.3) |
|
| Smoking
history |
|
| 0.006 |
| Never
smoker | 18 (32.7) | 87 (54.0) |
|
| Former
or current smoker | 37 (67.3) | 74 (46.0) |
|
| EGFR gene |
|
| <0.001 |
|
Wild-type | 43 (78.2) | 75 (46.6) |
|
|
Mutant | 12 (21.8) | 86 (53.4) |
|
| Pathological
subtype |
|
| <0.001 |
|
Lepidic | 9 (16.4) | 36 (22.4) |
|
|
Papillary | 27 (49.1) | 99 (61.5) |
|
|
Acinar | 5 (9.1) | 12 (7.5) |
|
|
Solid | 7 (12.7) | 10 (6.2) |
|
| Other
variants | 7 (12.7) | 4 (2.4) |
|
| Tumor size |
|
| 0.164 |
| ≤3
cm | 29 (52.7) | 102 (63.4) |
|
| >3
cm | 26 (47.3) | 59 (36.6) |
|
| LN metastasis |
|
| 0.706 |
| N0 | 46 (83.6) | 131 (81.4) |
|
|
N1/N2/N3 | 9 (16.4) | 30 (18.6) |
|
| Distant
metastasis |
|
| 1.000 |
| M0 | 54 (98.2) | 159 (98.8) |
|
| M1 | 1 (1.8) | 2 (1.2) |
|
| Pleural
invasion |
|
| 0.598 |
|
(−) | 39 (70.9) | 120 (74.5) |
|
|
(+) | 16 (29.1) | 39 (24.2) |
|
| Lymphatic
invasion |
|
| 0.184 |
|
(−) | 44 (80.0) | 114 (70.8) |
|
|
(+) | 11 (20.0) | 47(29.2) |
|
| Vascular
invasion |
|
| 0.217 |
|
(−) | 37 (67.3) | 122 (75.7) |
|
|
(+) | 18 (32.7) | 39 (24.2) |
|
 | Table II.Associations between
clinicopathological characteristics of lung adenocarcinoma patients
and FAM83B expression in tumors. |
Table II.
Associations between
clinicopathological characteristics of lung adenocarcinoma patients
and FAM83B expression in tumors.
| Variable | Odds ratio | 95% confidence
interval | P-value |
|---|
| Univariate
analysis |
|
|
|
| ≥70
years old | 1.411 | 0.763–2.609 | 0.272 |
|
Male | 2.468 | 1.277–4.769 | 0.007 |
|
Smoking | 2.417 | 1.271–4.596 | 0.007 |
|
Wild-type EGFR | 0.243 | 0.120–0.495 | <0.001 |
|
Histological Subtype | 2.202 | 0.795–6.101 | 0.129 |
| Tumor
size >3 cm | 1.550 | 0.835–2.878 | 0.165 |
| LN
metastasis | 0.854 | 0.377–1.934 | 0.706 |
| Distant
metastasis | 1.472 | 0.131–16.560 | 0.754 |
| Pleural
invasion | 1.201 | 0.607–2.373 | 0.599 |
|
Lymphatic invasion | 0.606 | 0.288–1.275 | 0.187 |
|
Vascular invasion | 1.522 | 0.780–2.970 | 0.218 |
| Multivariate
analysis |
|
|
|
|
Wild-type EGFR | 0.243 | 0.120–0.495 | <0.001 |
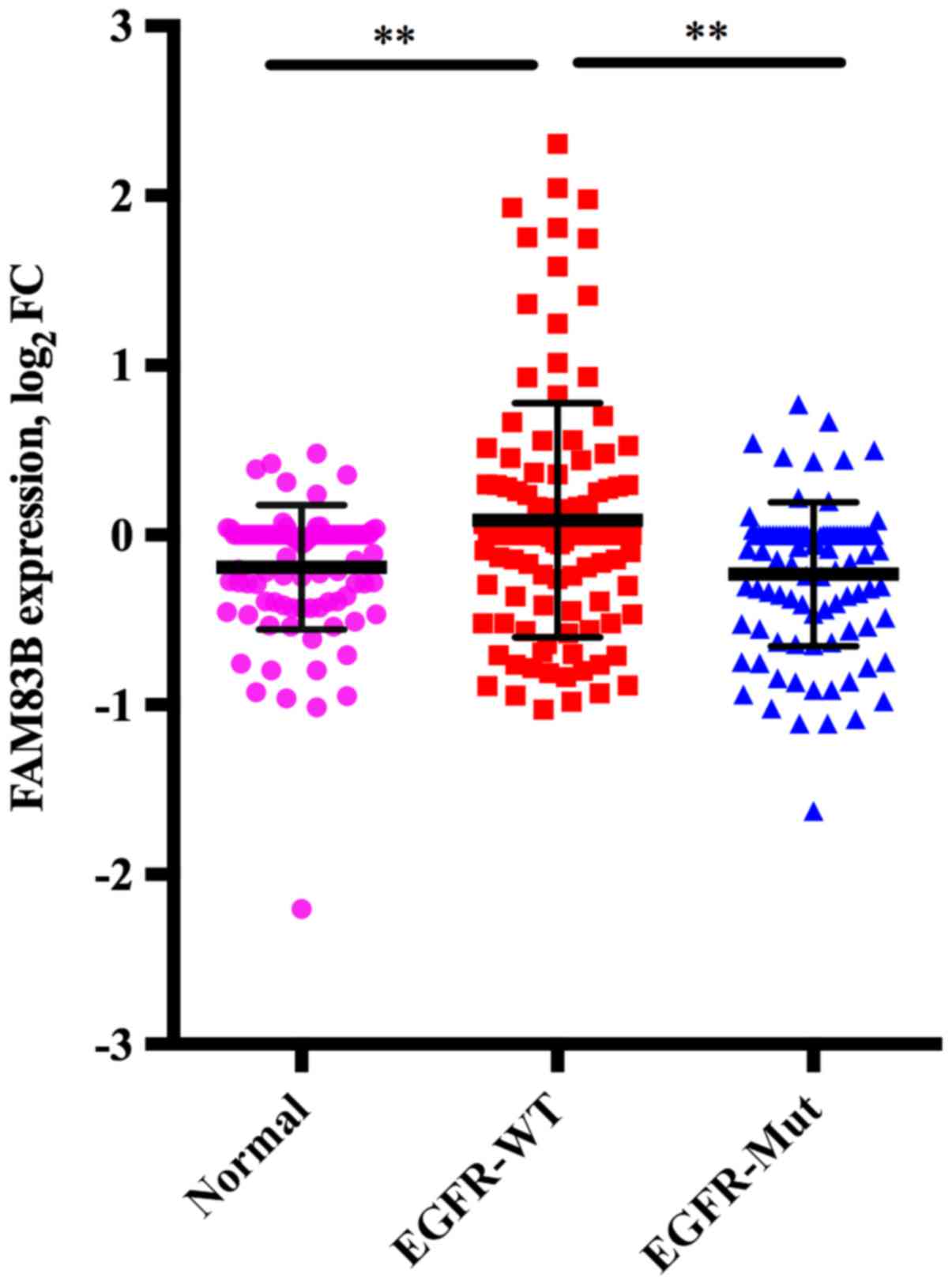
Correlation between FAM83B expression
obtained from cDNA microarray analysis and EGFR mutation in lung
ADC
The mean FAM83B expression in adjacent normal
lung tissue (n=98), wild-type EGFR tumors (n=118) and mutant
EGFR tumors (n=98) was −0.190, standard deviation (SD) of
0.365, 0.877, SD of 0.689, and −0.231, SD of 0.425, respectively.
Multiple comparison of these three groups showed that FAM83B
expression in wild-type EGFR tumors was higher than in
adjacent normal lung tissue (P<0.001) or mutant EGFR
tumors (P<0.001), while there was no significant difference
between mutant EGFR tumors and adjacent normal lung tissue
(P=0.852) (Fig. 1).
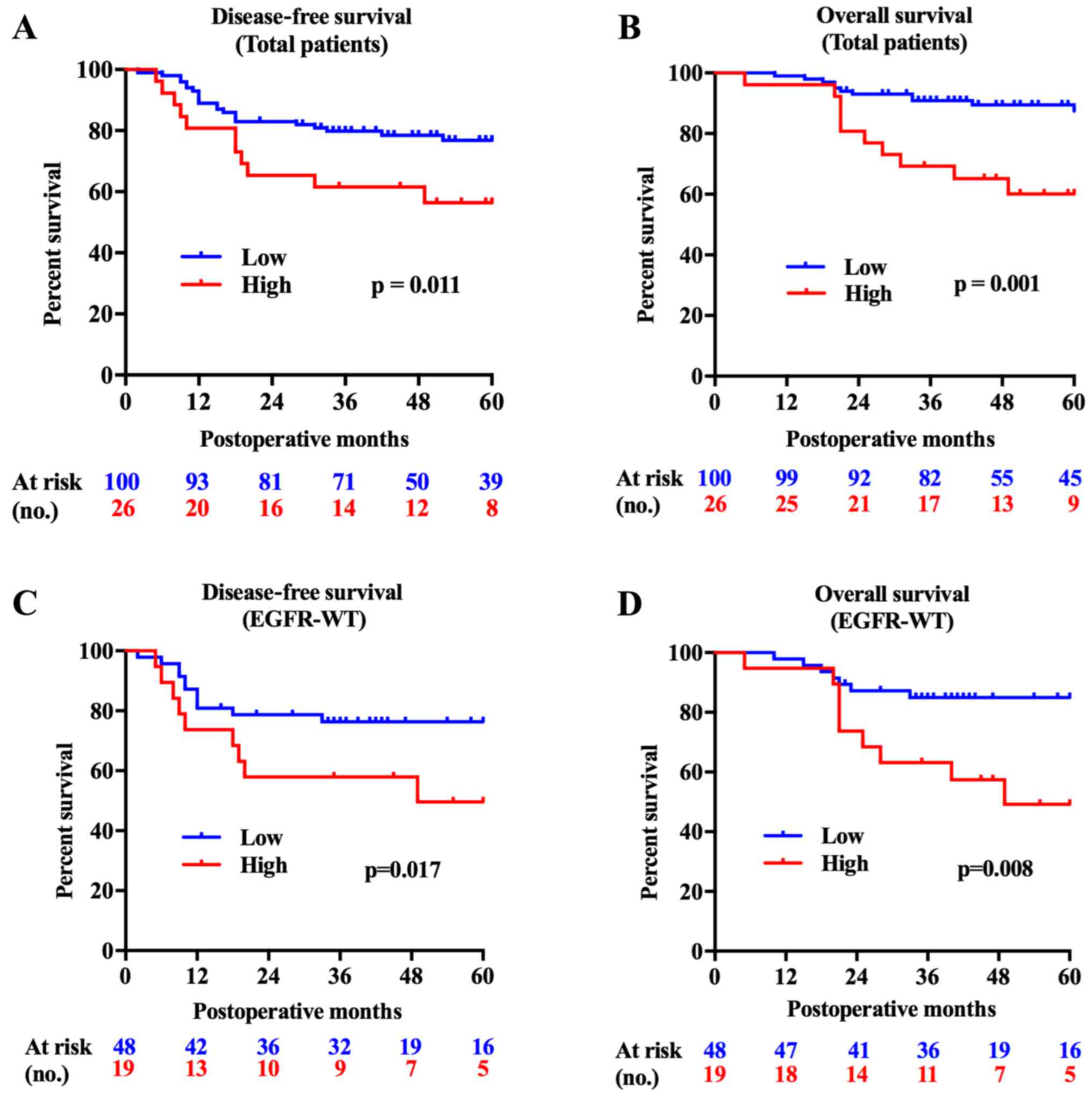
FAM83B is a predictor of poor lung ADC
prognosis, especially for ADC with wild-type EGFR
The FAM83B high expression group showed
significantly shorter survival times both in DFS (P=0.011) and OS
(P=0.001). Subgroup analysis showed that the FAM83B high
expression group had shorter DFS and OS with wild-type EGFR
tumors (P=0.017, P=0.008, Fig. 2),
while no significant difference was found in patients with mutant
EGFR tumors (P=0.746, P=0.588). Survival analysis using a
Cox regression hazard model was then conducted. For DFS, univariate
analysis showed that high levels of FAM83B expression, male
sex, lymph node metastasis, pleural invasion, lymphovascular
invasion, and vascular invasion were involved in poor prognosis.
Multivariate analysis identified high levels of FAM83B
expression and lymph node metastasis as independent poor prognostic
factors (Table III). In OS,
univariate analysis showed high levels of FAM83B expression,
male sex, wild-type EGFR tumors, lymph node metastasis,
pleural invasion, lymphovascular invasion, and vascular invasion as
poor prognostic factors. Multivariable analysis showed high levels
of FAM83B expression, pleural invasion, and vascular
invasion were independent predictors of poor prognosis (Table III).
 | Table III.Univariate and multivariate
predictors of DFS and OS. |
Table III.
Univariate and multivariate
predictors of DFS and OS.
| A, DFS |
|---|
| Variable |
Unfavorable/favorable | Hazard ratio (95%
confidence interval) | P-value |
|---|
| Univariate
analysis |
|
|
|
|
FAM83B | High/low | 2.415
(1.195–4.881) | 0.014 |
|
Age | ≥70/<70 | 1.183
(0.604–2.318) | 0.624 |
|
Sex | Male/female | 2.502
(1.196–5.233) | 0.015 |
| EGFR
gene |
Wild-type/mutant | 0.595
(0.298–1.189) | 0.142 |
|
Pack-year | >5/≤5 | 1.455
(0.735–2.881) | 0.282 |
| Tumor
size | >3 cm/≤3 cm | 1.073
(0.537–2.142) | 0.843 |
| N | N1+N2+N3/N0 | 3.852
(1.867–7.948) | <0.001 |
| pl |
Positive/negative | 2.599
(1.299–5.197) | 0.007 |
| ly |
Positive/negative | 2.347
(1.173–4.696) | 0.016 |
| v |
Positive/negative | 2.929
(1.477–5.811) | 0.002 |
| Multivariate
analysis |
|
|
|
|
FAM83B | High/low | 2.286
(1.129–4.631) | 0.022 |
| N | N1+N2+N3/N0 | 3.699
(1.788–7.655) | <0.001 |
|
| B, OS |
|
|
Variable |
Unfavorable/favorable | Hazard ratio
(95% confidence interval) | P-value |
|
| Univariate
analysis |
|
|
|
|
FAM83B | High/low | 3.814
(1.619–8.989) | 0.002 |
|
Age | ≥70/<70 | 1.010
(0.429–2.380) | 0.981 |
|
Sex | Male/female | 3.241
(1.187–8.854) | 0.022 |
| EGFR
gene |
Wild-type/mutant | 0.297
(0.108–0.813) | 0.018 |
|
Pack-year | >5/≤5 | 2.080
(0.839–5.154) | 0.114 |
| Tumor
size | >3 cm/≤3 cm | 1.297
(0.547–3.079) | 0.555 |
| N | N1+N2+N3/N0 | 4.342
(1.798–10.483) | 0.001 |
| pl |
Positive/negative | 5.313
(2.234–12.634) | <0.001 |
| ly |
Positive/negative | 2.577
(1.085–6.117) | 0.032 |
| v |
Positive/negative | 3.338
(1.414–7.877) | 0.006 |
| Multivariate
analysis |
|
|
|
|
FAM83B | High/low | 3.723
(1.568–8.842) | 0.003 |
| pl |
Positive/negative | 5.098
(2.119–12.264) | <0.001 |
| v |
Positive/negative | 2.529
(1.066–6.001) | 0.035 |
Involvement of FAM83B in cell
proliferation in several types of lung cancer cell line
Cell lines derived from lung ADC, including HLC-1,
H2347 (both wild-type for EGFR) and H1975 (mutant
EGFR), and a positive control breast cancer cell line, MCF7
(15) were prepared. RT-qPCR showed
high levels of FAM83B expression in HLC-1 and H2347 cells
but scarcely detectable levels in H1975 cells (Fig. 3A). Immunoblot analysis showed levels
of FAM83B that were consistent with the HLC-1, MCF7, and H1975
RT-qPCR results (Fig. 3B). In MCF7
cells, FAM83B knockdown with siRNA-8 or siRNA-9 caused
inhibition of cell proliferation (siRNA-8; P=0.393, siRNA-9;
P=0.061 at 6 days after knockdown), with siRNA-9 having the
stronger anti-proliferative effect (Fig.
4E-G); therefore, we performed subsequent knockdown experiments
using siRNA-9. siRNA transfection efficacy assays (Fig. 4A-D) indicated FAM83B RNAi
should be performed in HLC-1 and H1975 cells. Depletion of
FAM83B expression and suppression of cell proliferation were
confirmed in HLC-1 and even in H1975 cells, which expressed low
levels of FAM83B (Fig.
3C-H).
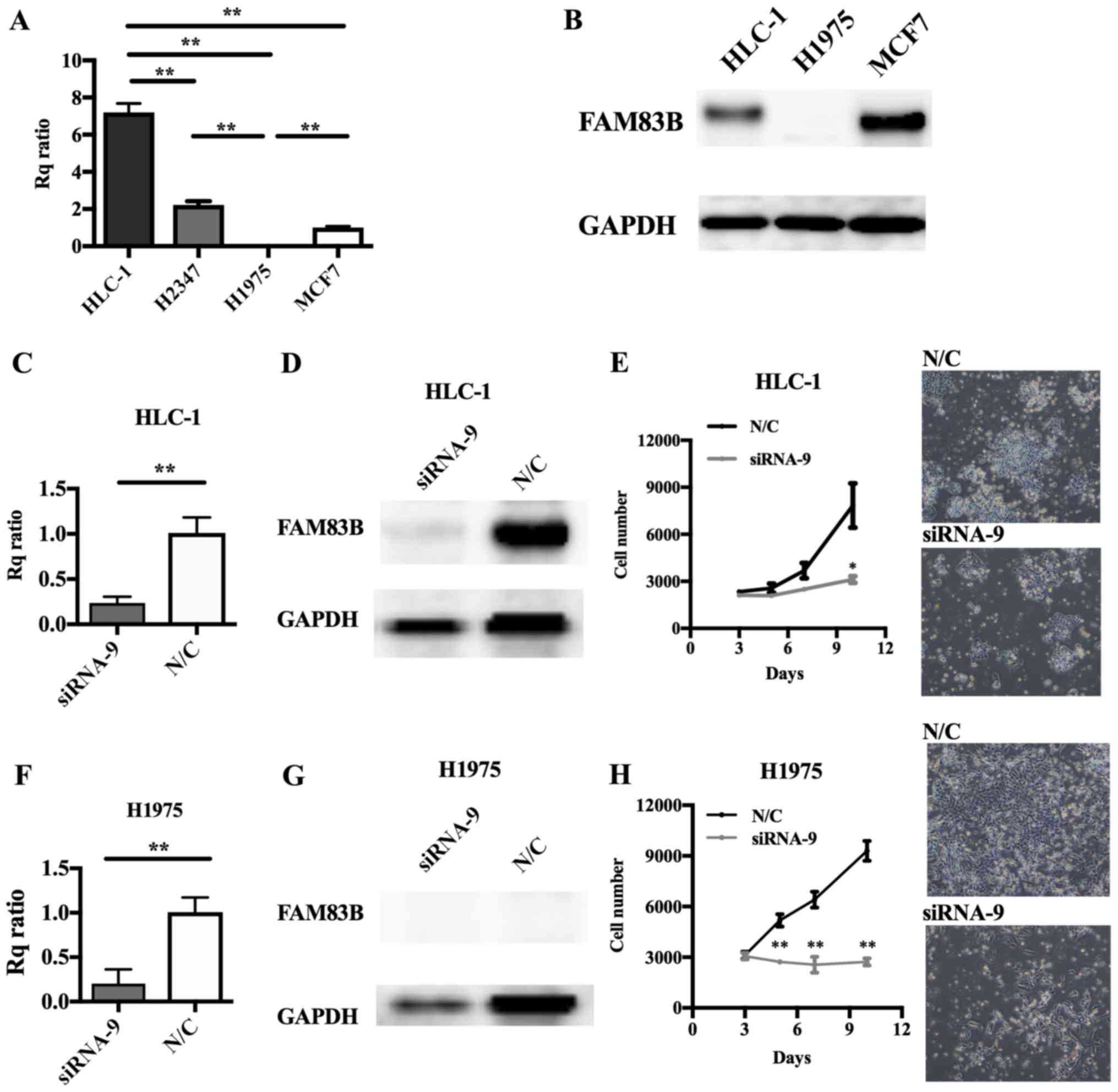 | Figure 3.Effect of FAM83B on the
proliferation of lung cancer cell lines. (A) FAM83B mRNA and
(B) protein levels were examined by RT-qPCR and western blotting,
respectively, in MCF7, H1975, H2347 and HLC1 cells. The Rq ratios
for RT-qPCR to MCF7 cells were as follows: H1975, 0.0061; H2347,
2.2103; and HLC1, 7.1968. (C-H) RNAi was performed in HLC-1 and
H1975 cells. Depletion of FAM83B mRNA and protein was
confirmed by (C and F) RT-qPCR and (D and G) western blotting,
respectively. Rq ratios to negative control were as follows: HLC-1,
0.3519; and H1975, 0.1675 for RT-qPCR. **P<0.01, as indicated.
Cell proliferation assay in (E) HLC-1 and (H) H1975 cells following
RNAi. Cell numbers were counted over time. Cell growth was
significantly suppressed in HLC-1 and H1975 cells. Magnification,
×40. Data are presented as the mean ± standard deviation.
*P<0.05 and **P<0.01 vs. N/C. FAM83B, family with sequence
similarity 83, member B; Rq ratio, relative quantification ratio;
N/C, negative control; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; RNAi, RNA interference. |
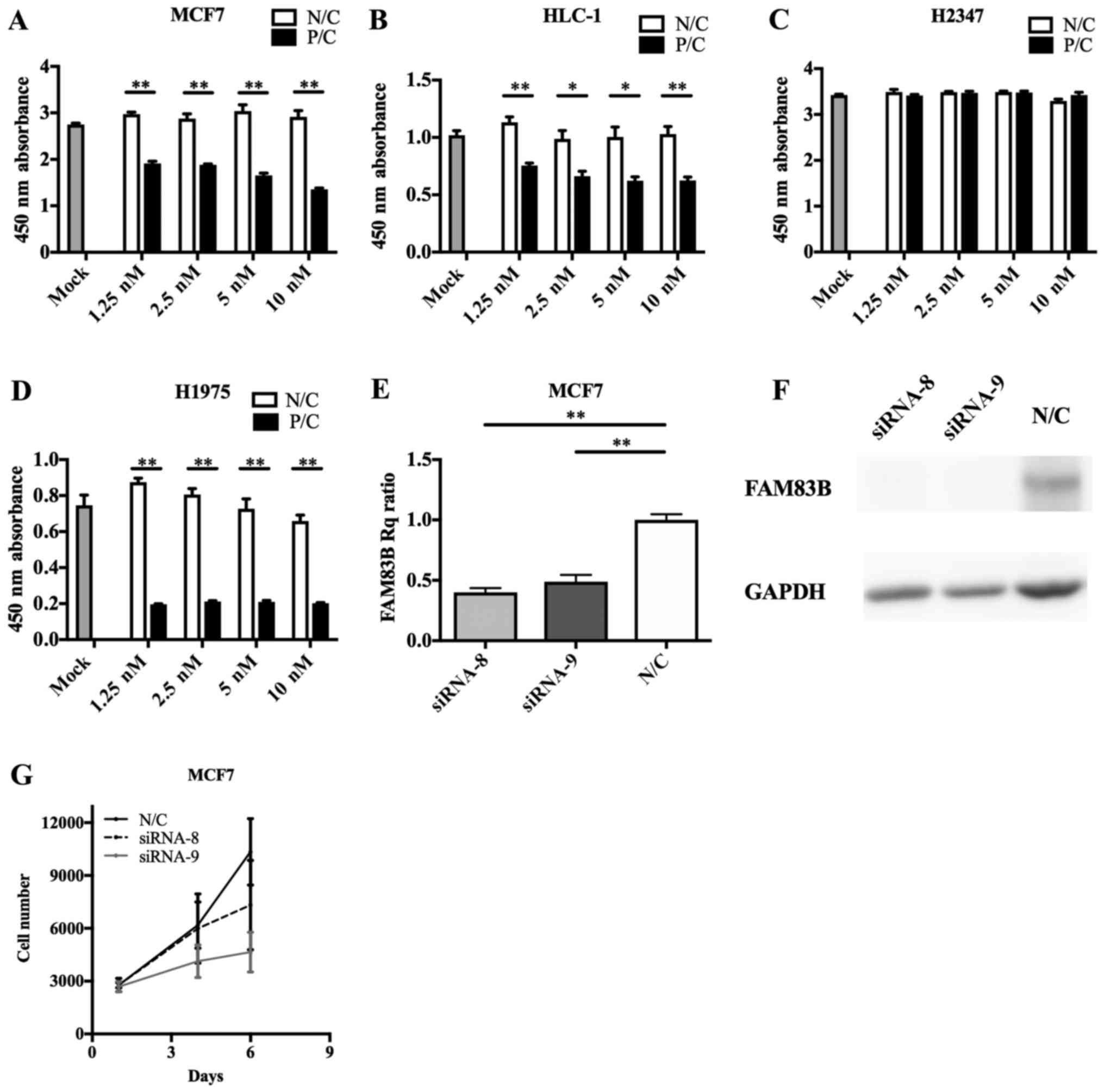 | Figure 4.FAM83B siRNA transfection
efficiency and RNA interference in cultured cells. Transfection of
FAM83B siRNA significantly suppressed the cell growth of (A)
MCF7, (B) HLC-1 and (D) H1975 cells, though not that of (C) H2347
cells. (E) Reverse transcription-quantitative polymerase chain
reaction revealed that siRNA-8 and siRNA-9 reduced the expression
of FAM83B. (F) Immunoblotting analysis showed that FAM83B
signals were markedly depleted in MCF7 cells transfected with
siRNA-8 or siRNA-9. (G) siRNA-8 and siRNA-9 suppressed cell growth,
particularly that of siRNA-9. Data are presented as the mean ±
standard deviation. *P<0.05 and **P<0.01, as indicated.
FAM83B, family with sequence similarity 83, member B; siRNA, small
interfering RNA; Rq, quantification relative to negative control;
P/C, positive control; N/C, negative control. |
Discussion
In this study, we focused on comprehensive gene
expression analysis of tumors from resected lungs of Japanese ADC
patients, and we showed that FAM83B expression was higher in
tumors with wild-type EGFR compared with tumors with mutant
EGFR, and that FAM83B expression was associated with
proliferation in cell lines. Furthermore, FAM83B expression
was identified as a biomarker of poor prognosis from patient
clinical outcomes.
FAM83B was first reported by Cipriano and
colleagues (15) to be correlated
with anchorage-independent growth in breast cancer cell lines using
a validation-based insertion mutagenesis method. FAM83B is a
member of the FAM83 family, which includes eight members
(FAM83A to H) characterized by a domain of unknown
function 1669 (DUF1669), which contains an N terminal phospholipase
D-like motif. DUF1669 is considered to be involved in tumor
proliferation and oncogenesis (15).
FAM83B expression is elevated relative to normal associated
tissues in several types of cancer, such as breast, ovary,
cervical, testis, thyroid, bladder, lymphoid and lung (15). In lung cancer, analysis of
FAM83B expression confirmed that FAM83B expression
was significantly elevated in tumor specimens relative to normal
tissues (15). It was also reported
that FAM83B mRNA levels were significantly higher in SqCC
than in normal lung or ADC (14). It
is a novel finding that FAM83B is more highly expressed in
lung ADCs containing wild-type EGFR compared with ADCs
carrying mutant EGFR. Interestingly, in breast cancer,
FAM83B is expressed at higher levels in tumors without
estrogen and progesterone receptors or human EGFR-2, compared with
receptor-positive tumors (21).
Furthermore, FAM83H, the paralog of FAM83B, is
overexpressed in androgen independent-prostate cancer (22). These findings could indicate that
FAM83B has an oncogenic role without aberrant signals from
driver gene mutations or overexpressed hormone receptors. The
function of FAM83B remains unclear; however, a bifunctional
interaction mechanism with responses to EGF was proposed. EGF
signals to multiple growth and survival pathways, including the
RAS/RAF/MEK/ERK and the PI3K/AKT pathways. FAM83B may
interfere with the binding of 14-3-3 protein to CRAF, thereby
promoting membrane localization of CRAF, and promoting downstream
signals to MAPK (15,23,24).
Furthermore, FAM83B may also bind to p85 and p110 subunits
of PI3K to promote PI3K/AKT signaling (25), which promotes oncogenic transformation
through phospholipase D activation (26), and shows resistance against EGFR-TKI
(15,24,27). Our
knockdown study of FAM83B showed that its expression was
associated with tumor proliferation in cell lines expressing high
levels of FAM83B; however, H1975 cells showed that knockdown
of low levels of FAM83B also inhibited proliferation. Of all
the FAM83 members, only FAM83H knockout mice, which
live for only 2 weeks, have been reported, and FAM83H is
proposed to play a role in the maintenance of cell homeostasis
(22,28). Other FAM83 family members, including
FAM83B, may also be required at certain levels, even in
normal cells.
FAM83 family members are usually associated
with poor cancer prognosis (21). Our
study of lung ADC also showed that FAM83B correlates with
poor prognosis; however, our previous study reported high
FAM83B expression to be a biomarker for good DFS prognosis
in lung SqCC (14). Snijders et
al conducted a meta-analysis of several databases (29–31) and
suggested that lung ADC expresses relatively high levels of
FAM83A, D, E, F, and H, but FAM83A, B, D, and
F correlate with prognosis. In lung SqCC, all FAM83
members except FAM83E are highly expressed, but only
FAM83A correlates with poor prognosis (21). These findings indicate FAM83B
has separate roles among cancers or tumor subtypes.
FAM83 family members, including
FAM83B, are more highly expressed in lung cancer than in
normal lung tissue, and their expression is reflected in the T
factor of TNM classification (20,29,32). Our
study did not show a correlation between tumor size and
FAM83B expression. This contradiction was caused by the fact
that the T factor includes not only tumor size but pleural
invasion. Both tumor size and pleural invasion had no significant
effect on FAM83B expression.
FAM83B RNAi suppressed proliferation of human
lung cancer cell lines. This result indicates that FAM83B
could be a potential therapeutic target against EGFR-WT
malignancies, which account for 47–88.7% of lung ADC (4,29) and
which are rarely indicated for molecular targeted therapies.
Limitations of this study include lack of evaluation of advanced or
recurrent cancer cohorts because the patient cohort was derived
from operable lung ADC cases biased to early stages. Moreover, this
study did not examine other driver mutations of lung ADC, such as
KRAS, ALK fusions. Subdivision based on other genomic or molecular
features could further explain the function of FAM83B in
lung ADC. Further investigation is required.
Acknowledgements
This study was partially supported by a grant for
translational research programs of Fukushima Prefecture. H. Suzuki
received research support from Bristol Myers Squibb, AstraZeneca,
and Tsumura, outside the submitted study. T. Yamaura received
research support from AstraZeneca, outside the submitted study. S.
Muto had been employed by AstraZeneca, outside the submitted study.
Y. Yanagisawa had been employed by Nippon Gene Co., Ltd., outside
the submitted study. R. Honma is employed by Nippon Gene Co., Ltd.,
outside the submitted study. The authors thank H I, M Otsuki, E
Otomo, and Y Kikuta for their technical supports.
References
|
1
|
Stewart BW and Wild CP: World Cancer
Report 2014. IARC Press; Lyon: 2014, http://publications.iarc.fr/Non-Series-Publications/World-Cancer-Reports/World-Cancer-Report-2014
|
|
2
|
National Cancer Institute, . https://seer.cancer.govOctober 17–2003
|
|
3
|
National Comprehensive Cancer Network, .
NCCN Clinical Practice Guidelines in Oncology. https://www.nccn.org/professionals/physician_gls/f_guidelines.aspApril
17–2003
|
|
4
|
Kohno T, Tsuta K, Tsuchihara K, Nakaoku T,
Yoh K and Goto K: RET fusion gene: Translation to personalized lung
cancer therapy. Cancer Sci. 104:1396–1400. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rosell R, Ichinose Y, Taron M, Sarries C,
Queralt C, Mendez P, Sanchez JM, Nishiyama K, Moran T, Cirauqui B,
et al: Mutations in the tyrosine kinase domain of the EGFR gene
associated with gefitinib response in non-small-cell lung cancer.
Lung cancer. 50:25–33. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shaw AT and Solomon B: Targeting
anaplastic lymphoma kinase in lung cancer. Clin Cancer Res.
17:2081–2086. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mok TS, Wu YL, Thongprasert S, Yang CH,
Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ishinose Y, et
al: Gefitinib or carboplatin-paclitaxel in pulmonary
adenocarcinoma. N Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa
K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, et al:
First-line crizotinib versus chemotherapy in ALK-positive lung
cancer. N Engl J Med. 371:2167–2177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Borghaei H, Paz-Ares L, Horn L, Spigel DR,
Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al:
Nivolumab versus docetaxel in advanced nonsquamous non-small-cell
lung cancer. N Engl J Med. 373:1627–1639. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Patel JD, Socinski MA, Garon EB, Reynolds
CH, Spigel DR, Olsen MR, Hermann RC, Jotte RM, Beck T, Richards DA,
et al: PointBreak: A randomized phase III study of pemetrexed plus
carboplatin and bevacizumab followed by maintenance pemetrexed and
bevacizumab versus paclitaxel plus carboplatin and bevacizumab
followed by maintenance bevacizumab in patients with stage IIIB or
IV nonsquamous non-small-cell lung cancer. J Clin Oncol.
31:4349–4357. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Park K, Tan EH, O'Byrne K, Zhang L, Boyer
M, Mok T, Hirsh V, Yang JC, Lee KH, Lu S, et al: Afatinib versus
gefitinib as first-line treatment of patients with EGFR
mutation-positive non-small-cell lung cancer (LUX-Lung 7): A phase
2B, open-label, randomised controlled trial. Lancet Oncol.
17:577–589. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chan BA and Hughes BG: Targeted therapy
for non-small cell lung cancer: Current standards and the promise
of the future. Transl Lung Cancer Res. 4:36–54. 2015.PubMed/NCBI
|
|
13
|
Sholl LM, Aisner DL, Varella-Garcia M,
Berry LD, Dias-Santagata D, Wistuba II, Chen H, Fujimoto J, Kugler
K, Franklin WA, et al: Multi-institutional oncogenic driver
mutation analysis in lung adenocarcinoma: The lung cancer mutation
consortium experience. J Thorac Oncol. 10:768–777. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Okabe N, Ezaki J, Yamaura T, Muto S, Osugi
J, Tamura H, Imai J, Ito E, Yanagisawa Y, Honma R, et al: FAM83B is
a novel biomarker for diagnosis and prognosis of lung squamous cell
carcinoma. Int J Oncol. 46:999–1006. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cipriano R, Graham J, Miskimen KL, Bryson
BL, Bruntz RC, Scott SA, Brown HA, Stark GR and Jackson MW: FAM83B
mediates EGFR- and RAS-driven oncogenic transformation. J Clin
Invest. 122:3197–3210. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Miura A, Honma R, Togashi T, Yanagisawa Y,
Ito E, Imai J, Goshima N, Watanabe S and Nomura N: Differential
responses of normal human coronary artery endothelial cells against
multiple cytokines comparatively assessed by gene expression
profiles. FEBS Lett. 580:6871–6879. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Laemmli UK: Cleavage of structural
proteins during the assembly of the head of bacteriophage T4.
Nature. 227:680–685. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Towbin H, Staehelin T and Gordon J:
Electrophoretic transfer of proteins from polyacrylamide gels to
nitrocellulose sheets: procedure and some applications. Proc Natl
Acad Sci USA. 76:pp. 4350–4354. 1979; View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Leslie HS, Mary KG and Christian W: TNM
classification of malignant tumours. 7th. Wiley-Blackwell; Oxford:
2009
|
|
21
|
Snijders AM, Lee SY, Hang B, Hao W,
Bissell MJ and Mao JH: FAM83 family oncogenes are broadly involved
in human cancers: An integrative multi-omics approach. Mol Oncol.
11:167–179. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bartel CA, Parameswaran N, Cipriano R and
Jackson MW: FAM83 proteins: Fostering new interactions to drive
oncogenic signaling and therapeutic resistance. Oncotarget.
7:52597–52612. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tzivion G, Luo Z and Avruch J: A dimeric
14-3-3 protein is an essential cofactor for Raf kinase activity.
Nature. 394:88–92. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cipriano R, Miskimen KL, Bryson BL, Foy
CR, Bartel CA and Jackson MW: Conserved oncogenic behavior of the
FAM83 family regulates MAPK signaling in human cancer. Mol Cancer
Res. 12:1156–1165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cipriano R, Miskimen KL, Bryson BL, Foy
CR, Bartel CA and Jackson MW: FAM83B-mediated activation of
PI3K/AKT and MAPK signaling cooperates to promote epithelial cell
transformation and resistance to targeted therapies. Oncotarget.
4:729–738. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cipriano R, Bryson BL, Miskimen KL, Bartel
CA, Hernandez-Sanchez W, Bruntz RC, Scott SA, Lindsley CW, Brown HA
and Jackson MW: Hyperactivation of EGFR and downstream effector
phospholipase D1 by oncogenic FAM83B. Oncogene. 33:3298–3306. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Grant S: FAM83A and FAM83B: Candidate
oncogenes and TKI resistance mediators. J Clin Invest.
122:3048–3051. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang SK, Hu Y, Yang J, Smith CE,
Richardson AS, Yamakoshi Y, Lee YL, Seymen F, Koruyucu M, Gencay K,
et al: Fam83 h null mice support a neomorphic mechanism for human
ADHCAI. Mol Genet Genomic Med. 4:46–67. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carithers LJ and Moore HM: The
genotype-tissue expression (GTEx) project. Biopreserv Biobank.
13:307–308. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Melé M, Ferreira PG, Reverter F, DeLuca
DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD,
Sullivan TJ, et al: Human genomics. The human transcriptome across
tissues and individuals. Science. 348:660–665. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkami P, et al: Oncomine 3.0: Genes, pathways and
networks in a collection of 18,000 cancer gene expression profiles.
Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|