Introduction
Chronic inflammation, driving carcinogenic pathways,
increases the risk of developing cancer (1,2), such as
ulcerative colitis (UC)-associated colorectal cancer (3–6). UC
features chronic, relapsing, and debilitating idiopathic
inflammation of the colon mucosa and submucosa, and patients with
long-standing UC are at high risk of neoplastic development
(3). Endoscopy remains the
gold-standard method for the detection and quantification of UC
inflammation. UC-associated carcinomas are often difficult to
detect endoscopically and to discriminate from inflammatory
regenerative epithelium (7,8). Inflammation is assessed by measuring
C-reactive protein (CRP) levels (9),
erythrocyte sedimentation rate (10),
interleukin-6 (IL-6) levels (11),
and tumor necrosis factor alpha (TNF alpha) levels (12); these characteristics are all known as
biomarkers of chronic inflammation in colon cancer. However, these
inflammatory markers are not sufficient for estimating the risk of
colon carcinoma (13).
Similarly, fecal calprotectin reflects neutrophil
migration to the intestinal mucosa, which occurs during intestinal
inflammation (14). Fecal
calprotectin levels correlate with the severity of mucosal
inflammation (15–18). However, it specificity is low in
suspected inflammatory bowel disease (IBD) in children (19). Therefore, although a number of
inflammatory markers are available, these are still being
developed.
We previously reported that interactions between
epithelial cells and the stromal microenvironment in UC may play an
important role in the carcinogenic pathway (3,20,21), suggesting the existence of a mechanism
by which the interaction between epithelial cells and stromal cells
leads to interstitial fibrosis.
Thus, we sought to identify proteins that
characterize the inflammatory microenvironment. In the present
study, using proteomics, we identified proteins differentially
expressed in active and inactive UC mucosal biopsies. Agarose
two-dimensional gel electrophoresis (agarose-2DE) and
matrix-assisted laser desorption ionization-time of flight/time of
flight mass spectrometry (MALDI-TOF/TOF MS) identified several
proteins possibly associated with the inflammatory colonic mucosa.
Immunoreactive staining of histological sections of surgically
removed samples using antibodies against these proteins revealed
peroxiredoxin 1 (PRDX1) as a candidate protein overexpressed in
active UC biopsy samples.
Materials and methods
Biopsy samples for proteomic
analysis
The present study was approved by the Committee for
Kitasato University Medical Ethics Organization (KMEO), and
informed consent was obtained from all patients prior to the
beginning of the study (KMEO B08-01 and KMEO B01-15).
Biopsy samples were obtained at Kitasato East
University Hospital from two patients with active UC, two patients
with inactive UC, and four unrelated normal healthy controls.
Biopsies were taken from two sites in each case: The sigmoid colon
and rectum. The UC activity was histologically determined by Matts
score (22) of neighboring biopsy
samples using the following criteria: Biopsy of an active UC of
sigmoid colon, Matts 4; biopsy of an active UC of rectum, Matts 2;
biopsy of an inactive UC of sigmoid colon and rectum, Matts 1 for
both.
Histological examination of UC
regenerative mucosa and UC-associated neoplastic lesions
UC regenerative mucosa samples were collected from
UC cases in the pathological collection of Kitasato University
Hospital and Kitasato University East Hospital. The corresponding
clinicopathological data are summarized in Table I.
 | Table I.Clinicopathological data of the
patients who provided UC regenerative specimens. |
Table I.
Clinicopathological data of the
patients who provided UC regenerative specimens.
|
| Number of
cases | Age (years) (mean ±
s.d.) | Gender (M:F) | Number of
samples | Number of samples
classified by Matts score | Duration from the
onset of UC (years mean ± s.d.) |
|---|
| Normal control | 22 | 61.0±13.7 | 11:11 | 22 |
|
|
| UC cases | 35 | 41.8±15.2 | 15:20 | 100 | 5 (Matts 1) | 12.4±1.3 |
|
|
|
|
|
| 22 (Matts 2) | 7.0±4.7 |
|
|
|
|
|
| 18 (Matts 3) | 6.7±5.1 |
|
|
|
|
|
| 55 (Matts 4,5) | 5.5±5.1 |
UC-associated neoplastic lesion samples were
previously described (21).
Thirty-nine UC regenerative mucosae, six UC-associated low-grade
dysplasias (LGD), five high-grade dysplasias (HGD), five
UC-associated carcinomas (UCCA), and 22 normal control mucosae were
collected.
Proteomic analysis
Protein extraction, agarose-2DE, in-gel digestion,
and protein identification were performed as previously described
(23,24). Peptide mass fingerprinting (PMF) was
performed using the Autoflex III MALDI-TOF/TOF MS (Bruker
Corporation, Ettlingen, Germany). PMF and MS/MS spectra were
submitted to MASCOT (http://www.matrixscience.com/search_from_select.html)
for searching of the IPI human 20,081,114 database (74,049
sequences, 31,194,560 residues; www.ebi.ac.uk/IPI/Databases.html/) and identification
of the corresponding proteins.
Western blot analysis
Western blot analysis was performed as previously
described (21) using anti-PRDX1
(monoclonal, 1:1,000; Abnova, Taipei, Taiwan) and anti-β-actin
(monoclonal, 1:10,000; Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) antibodies. Proteins were extracted from sigmoid colon
biopsy samples (two from active UC, two from inactive UC, and three
from normal controls). A UC-associated cancer cell line (UCCA-24)
and a sporadic colorectal cancer cell line (KE43P) served as colon
cancer controls (25).
Immunohistochemical staining
Immunohistochemical staining was performed as
previously described (21) using
anti-PRDX1 antibodies (monoclonal, 1:200; Abnova) and anti-TRX
antibodies (polyclonal, 1:1,000; Abcam, Cambridge, MA, USA).
Sections for TRX staining were incubated in 10 mM citrate buffer
(pH 6.0) and boiled in a microwave oven for 15 min. After
incubation with Protein Block Serum-Free (DakoCytomation, Glostrup,
Denmark), sections were incubated overnight with anti-PRDX1 and
anti-TRX antibodies at 4°C.
The immunoreactivities of UC regenerative mucosal
crypts were evaluated at the upper and lower halves independently.
The staining intensities of PRDX1 in UC regenerative and normal
mucosal epithelia were determined semi-quantitatively (none, 0;
weak, 1; moderate, 2; intense, 3). The number of positive cells per
250-µm mucosal length (mainly histiocytes) was counted in UC
regenerative and normal mucosal stroma. TRX expression in UC
regenerative and normal mucosal crypts was determined as the number
of positive crypts/the total number of crypts. Immunoreactivity in
UC-associated neoplastic lesions was evaluated by using a
previously described scoring system (26), which combines staining intensity
(none, 0; weak, 1; moderate, 2; intense, 3) with percentages of
positive cells (0, <1%; 1, 1–25%; 2, 25–50%; 3, 50–75%; 4,
>75%).
Statistical analysis
All statistical tests were conducted using StatView
v5.0 (SAS Institute, Inc., Cary, NC, USA) or SPSS v16.0 for Windows
(IBM, Inc., Chicago, IL, USA). For comparison of more than three
groups, Kruskal-Wallis tests were performed with the Mann-Whitney
U-test as a post-hoc test for comparisons between two
groups. P<0.05 was considered to indicate a statistically
significant difference. The correlation between the Matts score and
TRX protein expression was tested using Spearman's rank correlation
coefficient.
Results
Proteomic analysis
Thirteen spots showed a more than 1.5-fold
difference between active and inactive UC in agarose 2-DE and were
digested with trypsin. MALDI-TOF/TOF MS identified nine proteins
showing higher expression in active than in inactive UC and four
proteins showing lower expression in active than in inactive UC;
these results are shown in Table
II.
 | Table II.Proteins in active ulcerative
colitis. |
Table II.
Proteins in active ulcerative
colitis.
| A, Upregulated
proteins in active ulcerative colitis |
|---|
|
|---|
|
|
| Active/inactive
fold |
|---|
|
|
|
|
|---|
| Spot no. | Protein name | Sigmoid | Rectum |
|---|
| 19 | CyclophilinB | 2.0 | 1.6 |
| 22 | Ig kappa chain
V–III | 2.1 | 1.3 |
| 3 | SOD2 superoxide
dismutase | 2.0 | 1.5 |
| 28 |
peroxiredoxin-1 | 1.7 | 1.0 |
| 21-1 | L-lactate
dehydrogenase | 1.5 | 1.6 |
| 4-1 | HSP 60
mitochondrial | 2.0 | 1.3 |
| 5 | ETFB Isoform 1 of
Electron transfer flavoprotein subunit beta | 2.5 | 2.4 |
| 15 | IGHV4-31 anti-RhD
monoclonal T125 gammal heavy chain | 1.6 | 1.0 |
| 34 | ENO1 Isoform
alpha-enolase of Alpha-enolase | 1.6 | 1.1 |
|
| B, Downregulated
proteins in active ulcerative colitis |
|
|
|
| Active/inactive
fold |
|
|
|
|
| Spot
no. | Protein
name | Sigmoid | Rectum |
|
| 7 | Galectin-4 | 0.5 | 0.5 |
| 1 | Cytokeratin 19 | 0.6 | 0.4 |
| 6 | Carbonic anhydrase
1 | 0.4 | 0.4 |
| 10 | Cytokeratin 19 | 0.3 | 0.5 |
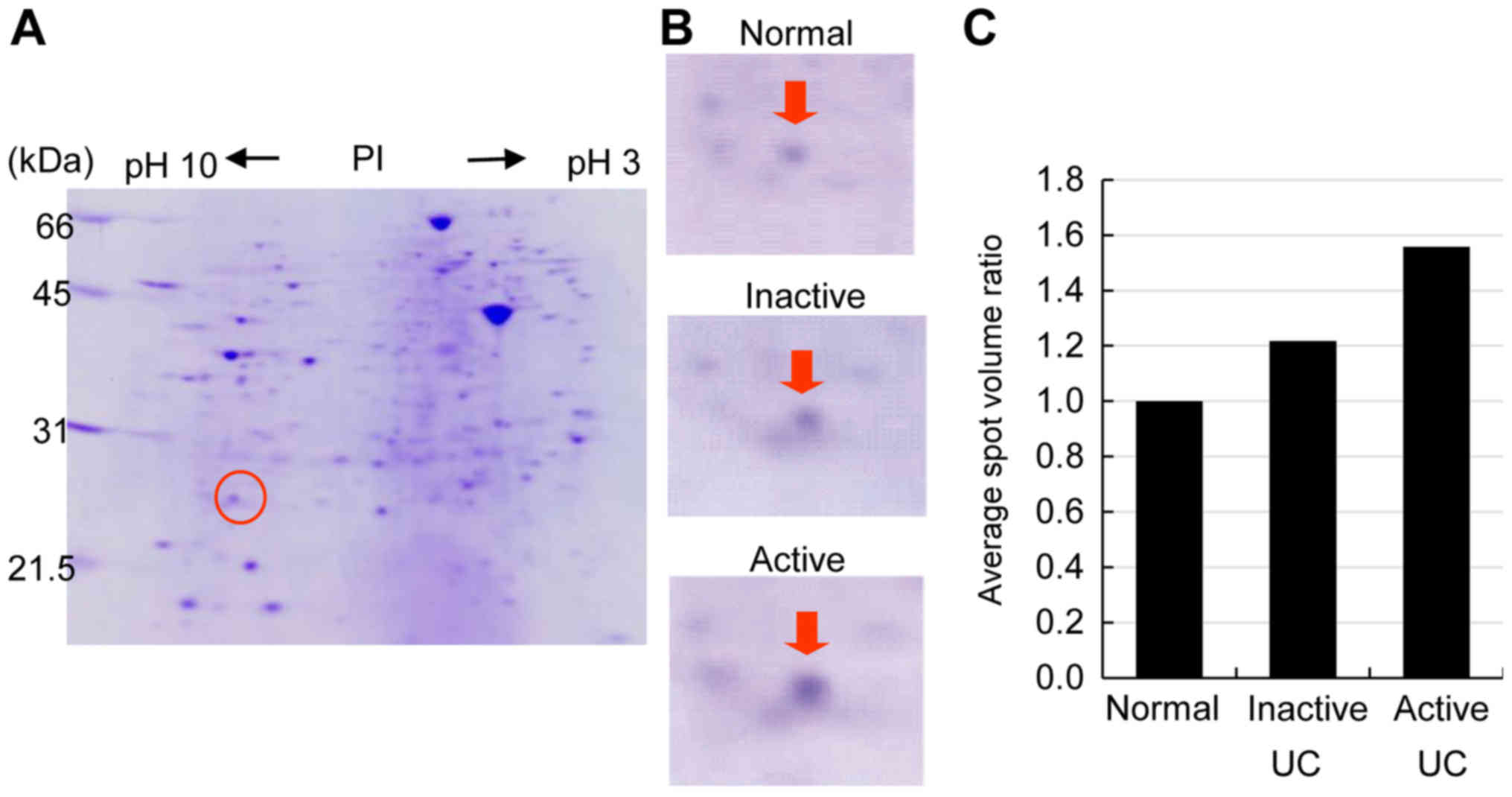
PRDX1 expression was consistently higher in active
than in inactive UC. PRDX1 spots from an active UC sigmoid colon
biopsy showed 1.7-fold higher expression than those from an
inactive UC sigmoid colon biopsy. A representative proteome map
derived from the active UC sigmoid colon biopsy specimen is shown
in Fig. 1A, while the relative ratio
of PRDX1 spot volumes is shown in Fig.
1C.
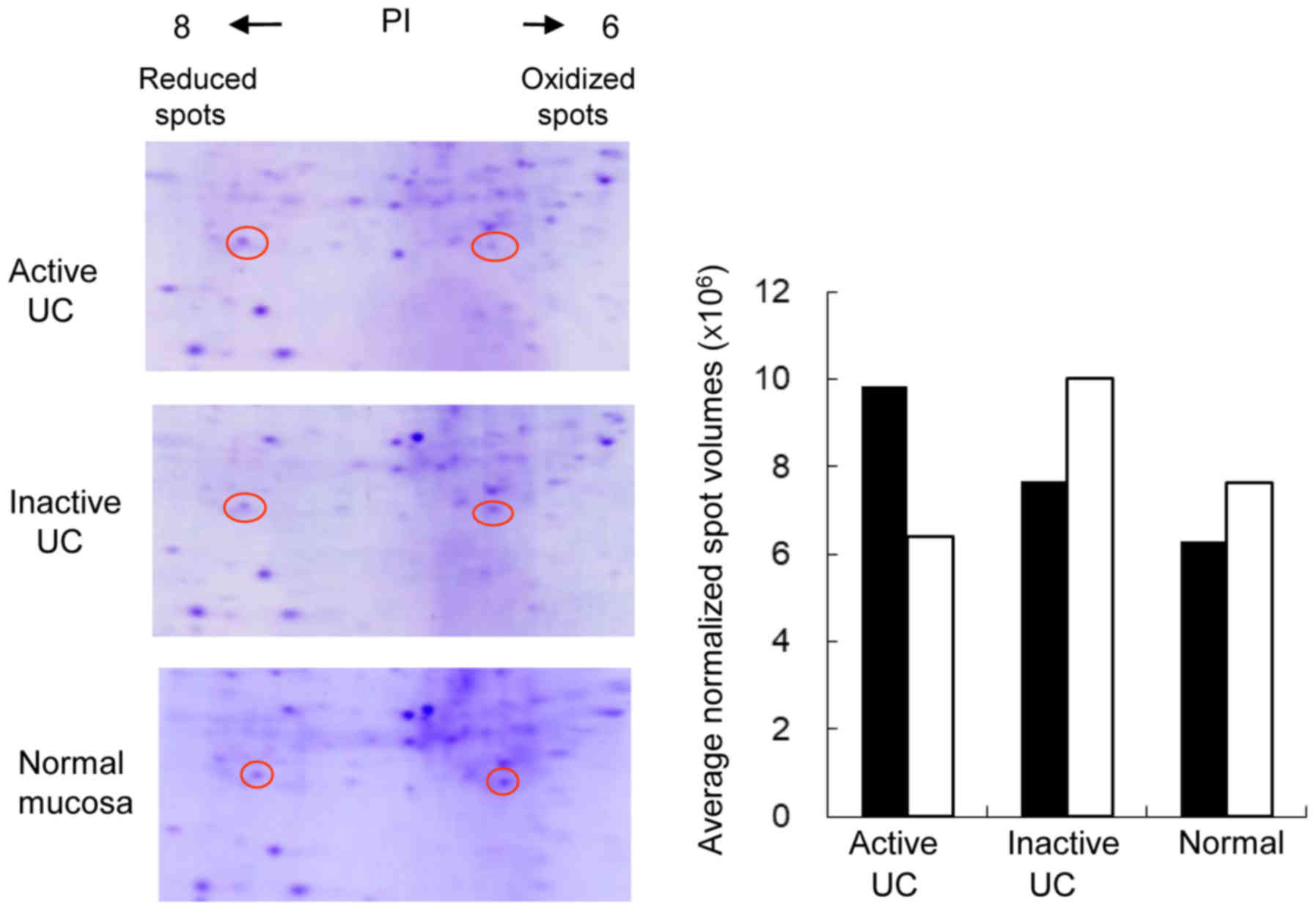
Two electrophoretic spots of PRDX1 with different
isoelectric points (PI) were identified using agarose-2DE. While
both of these were confirmed to be PRDX1, the spot with a PI around
6 reflected methionine oxidation according to MALDI-TOF/TOF MS
analysis. Thus, this spot represented the oxidized form of PRDX1,
while the spot with a PI of around 8 was the reduced form of PRDX1.
The reduced PRDX1 spots were bigger and darker than the oxidized
PRDX1 spots in active UC biopsy specimens. Conversely, oxidized
PRDX1 expression was higher in inactive UC and normal biopsy
specimens, as shown in Fig. 2.
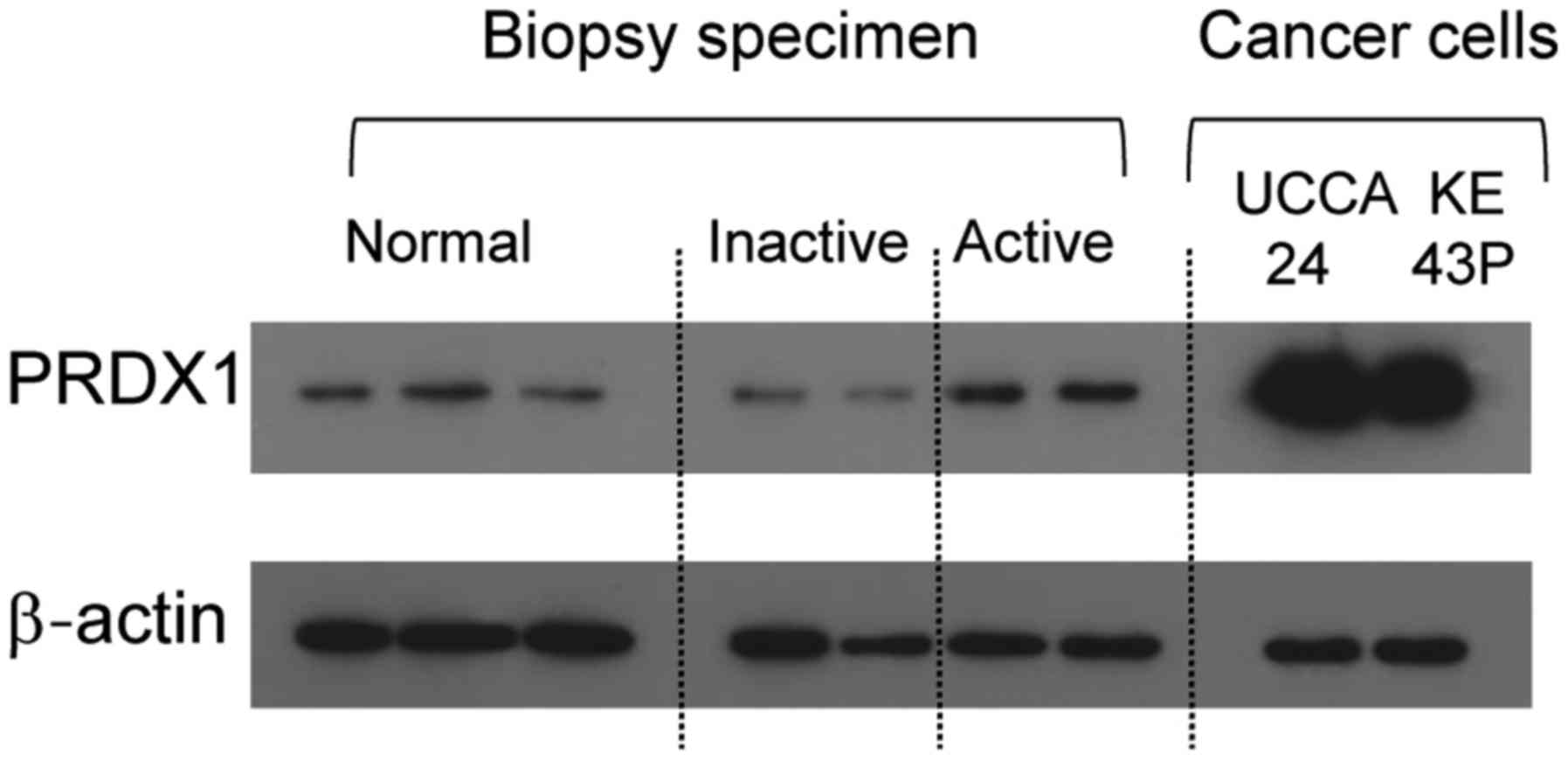
Western blot analysis of PRDX1
expression
As shown in Fig. 3,
PRDX1 expression was higher in active UC than in inactive UC and
normal colonic mucosae. In addition, PRDX1 was expressed in UCCA-24
and KE-43P colon cancer cell lines.
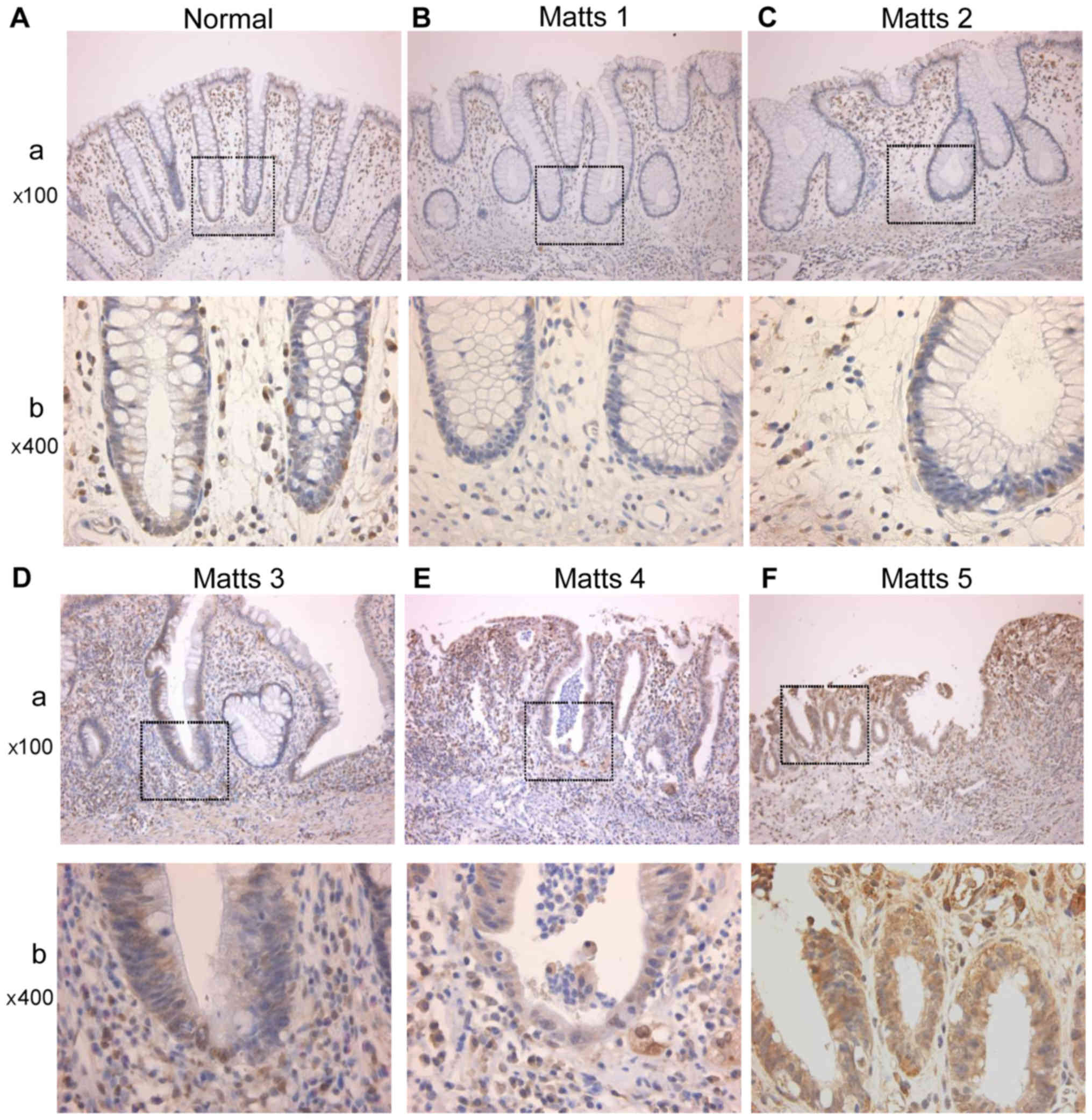
PRDX1 expression in UC regenerative
mucosae
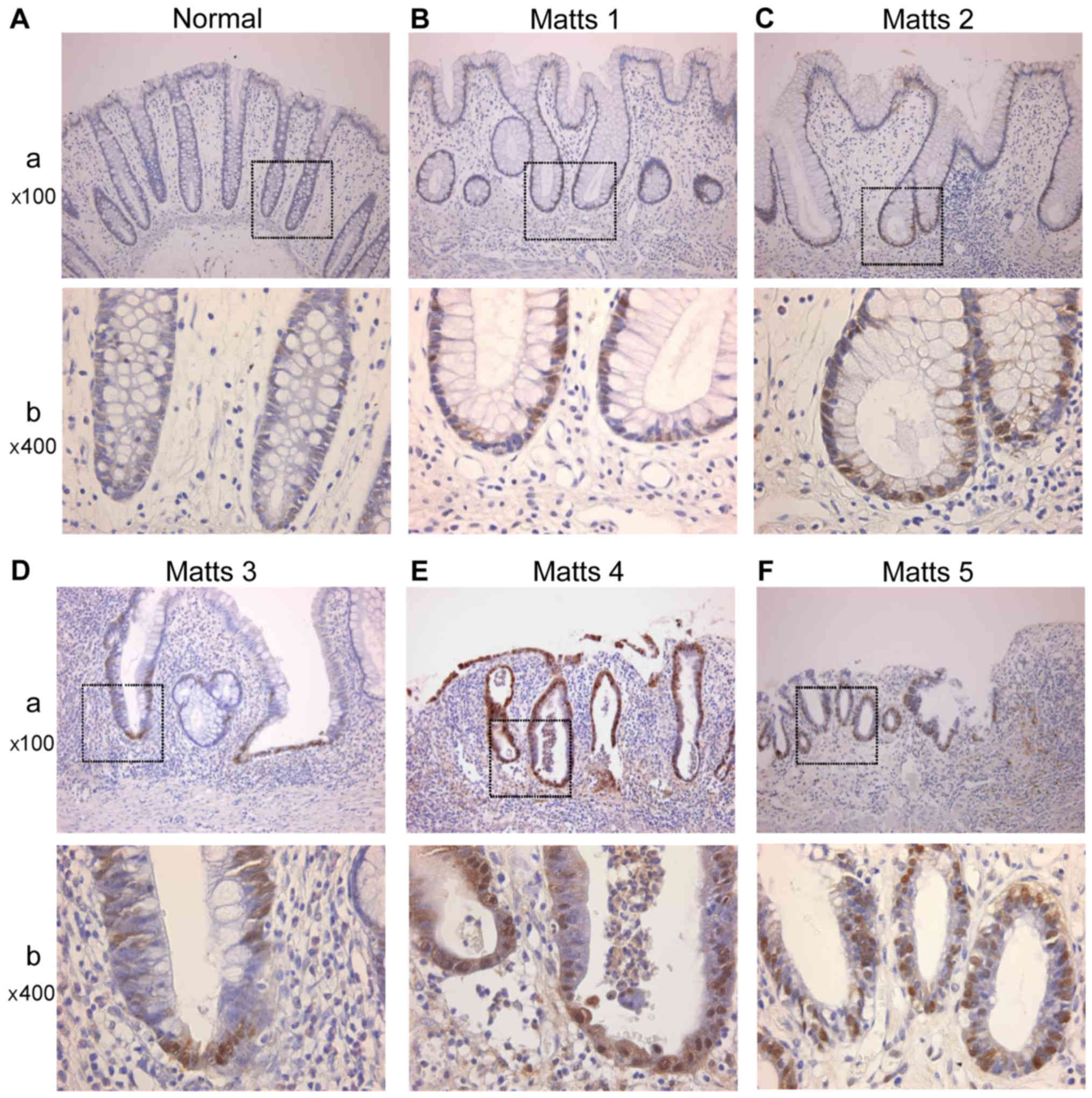
PRDX1 was expressed in the cytoplasm of epithelial
cells. In stromal tissue, PRDX1 was mainly expressed in
macrophages, as shown in Fig. 4.
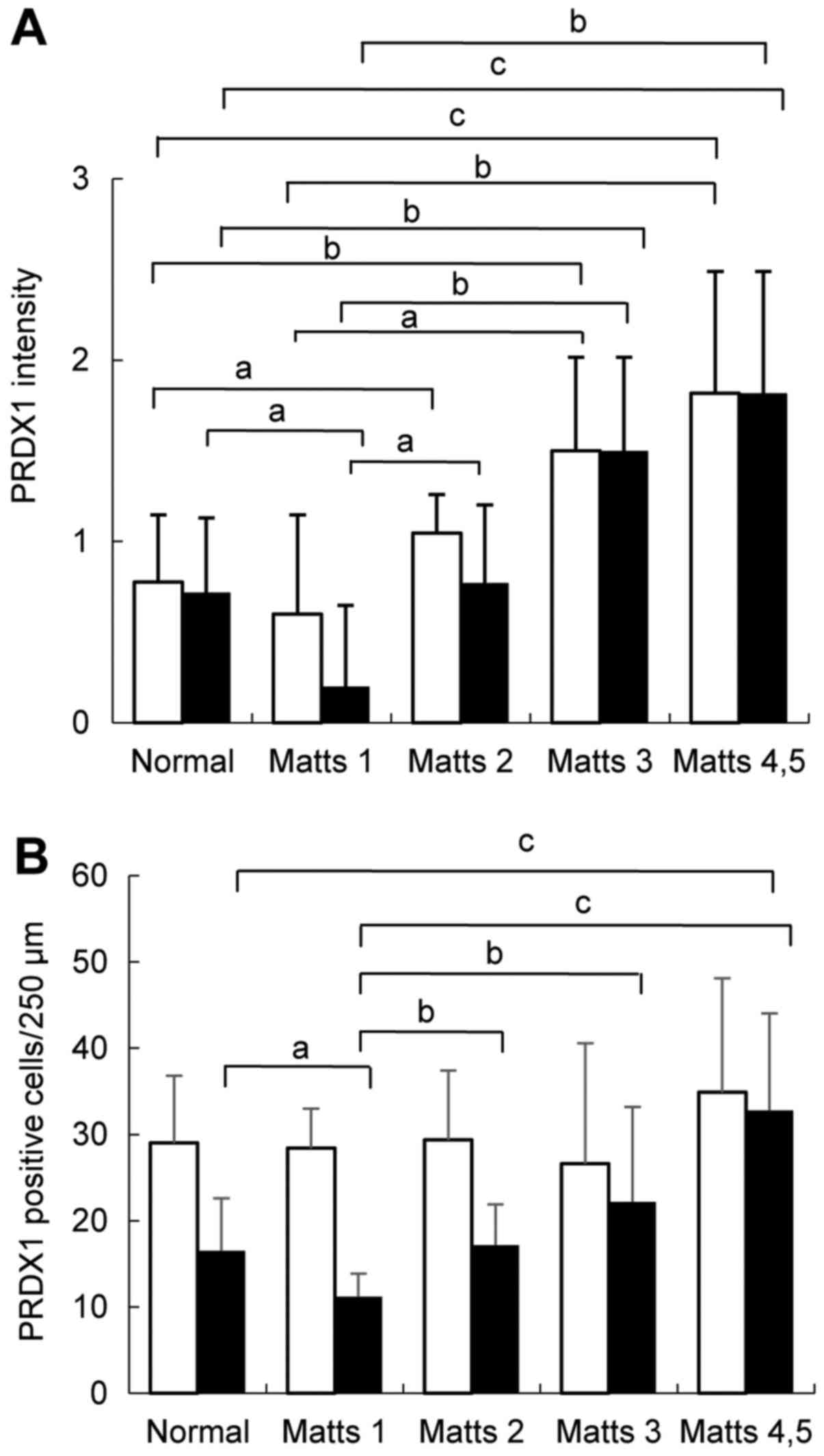
Fig. 5A shows PRDX1
immunohistochemical staining scores in UC regenerative and normal
colonic mucosae. Both the upper and lower halves of the crypts
showed increases in PRDX1 expression with increasing inflammation
levels as determined by Matts scores (Kruskal-Wallis test;
P<0.05). In particular, PRDX1 expression was significantly
higher in active UC than in the normal mucosa (Matts 3: P<0.01;
Matts 4–5: P<0.001; Fig. 5A). In
addition, Fig. 5B shows that in the
lower half of the stroma, the active UC mucosa exhibited a
significantly higher number of infiltrating PRDX1-positive cells
than did the normal mucosa (Matts 4–5: P<0.001). However, no
significant difference was observed in the upper half of the
stroma.
TRX expression in the UC-inflamed
colonic mucosa
TRX expression was mainly observed in the cytoplasm
of epithelial cells; however, some nuclear expression was also
observed. TRX expression was evident in the cytoplasm of epithelial
cells in active UC (Matts 3 and 4–5), while very weak in the normal
mucosae and inactive UC (Matts 1, 2). TRX expression tended to
increase gradually with increasing inflammation levels as
determined by Matts score in both the upper and lower halves of the
crypts, as shown in Fig. 6.
Crypts in which more than 50% of the epithelial
cells expressed TRX were considered TRX-positive. Ratios of
TRX-positive crypts (number of positive crypts/total number of
crypts) are summarized in Table
III. Ratios and Matts scores correlated in both the upper and
lower halves of the crypts; r=0.778 (P<0.001) for the
upper half, and r=0.821 (P<0.001) for the lower half.
 | Table III.TRX expression in mucosal crypts of
ulcerative colitis and normal colon. |
Table III.
TRX expression in mucosal crypts of
ulcerative colitis and normal colon.
|
| Normal (%) | Matts 1(%) | Matts 2 (%) | Matts 3 (%) | Matts 4,5 (%) |
|---|
| Upper half | 3/20 (15.0) | 0/5 (0) | 12/21 (57.1) | 14/15 (93.3) | 56/57a (98.2) |
| Lower half | 0/20 (0) | 1/5 (0.2) | 5/21 (23.8) | 12/15 (80.0) | 54/57b (94.7) |
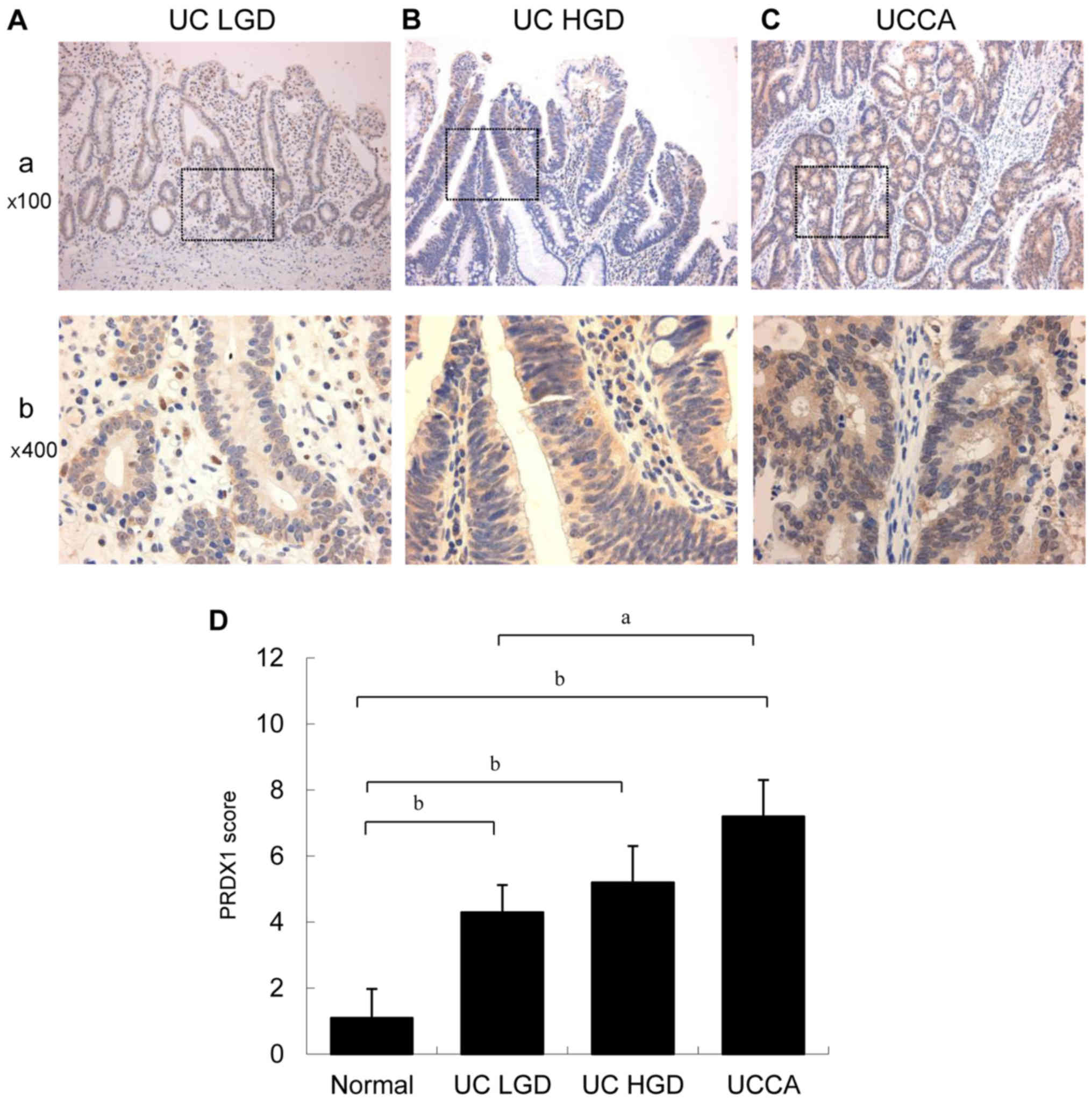
PRDX1 expression in UC-associated
dysplasia and carcinoma
Typical PRDX1 immunohistochemical staining patterns
of UC-associated neoplastic lesions are shown in Fig. 7A and B. PRDX1 expression gradually
increased from low-grade dysplasia to invasive carcinoma, as shown
in Fig. 7C. LGD, HGD, and UCCA showed
significantly higher PRDX1 expression than did the normal mucosa
(P<0.01).
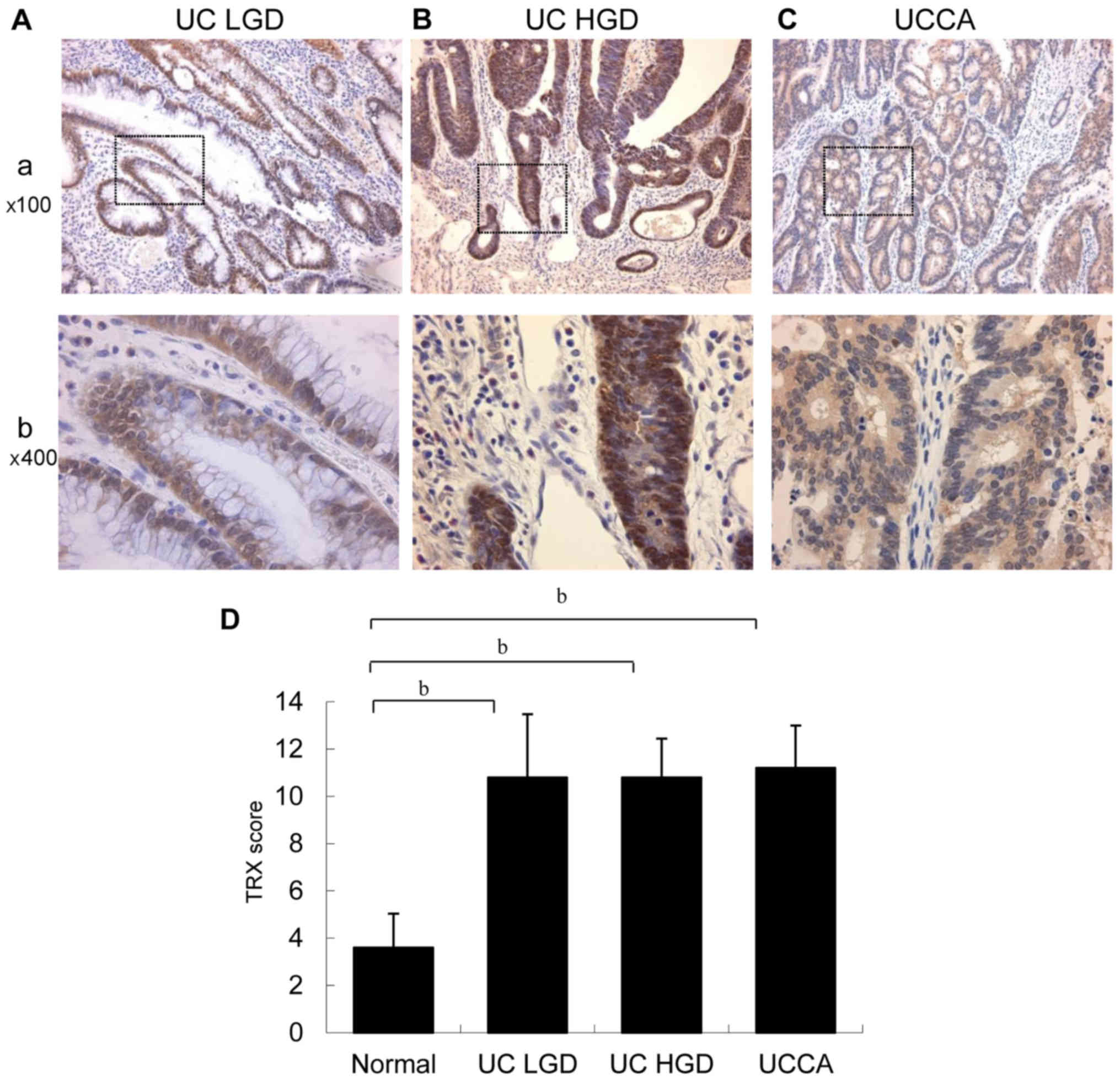
TRX expression in UC-associated
dysplasia and carcinoma
Typical TRX immunohistochemical staining patterns of
UC-associated neoplastic lesions are shown in Fig. 8A and B. TRX was mainly expressed in
the cytoplasm, but also partially observed in the nucleus in
UC-associated dysplasia and carcinoma. TRX expression in LGD, HGD,
and UCCA was significantly higher than that in the normal mucosa
(P<0.01), as shown in Fig. 8C.
However, no significant difference was observed among the types of
UC-associated neoplastic lesions (LGD, HGD, and UCCA).
Discussion
PRDXs are a class of thiol peroxidases that degrade
hydroperoxides to water (27). PRDXs
contain essential catalytic cysteine residues and are mainly
reduced by thioredoxins (TRXs) (28).
Mammals have six different PRDXs (PRDX1-6) (29). Various types of PRDXs have diverse and
even opposing functions (30). PRDX1
is a versatile molecule regulating cell growth, differentiation,
and apoptosis. PRDX1 is overexpressed in breast cancer (31), where it is significantly associated
with tumor invasion, nodal metastasis, and advanced disease stage
(32). Moreover, reduced PRDX1
expression is an important factor in esophageal squamous cancer
progression (33).
Oxidative stress plays an important role in the
carcinogenesis of various cancers (34). PRDX1 regulates ROS-dependent signaling
pathways, which play important roles in the progression and
metastasis of human tumors (35).
However, the biological function of PRDX1 in chronic
inflammation-induced carcinogenesis, such as in UC-associated
carcinoma, remains unclear (36).
In the present study, active and inactive UC biopsy
samples were compared using proteomics and immunohistochemistry.
PRDX1 was identified as an upregulated protein in active UC
specimens. PRDX1 expression increased with inflammation levels in
epithelial cells in the crypts of the UC regenerative mucosa.
Furthermore, PRDX1 was mainly expressed in macrophages in the
stroma of the UC regenerative mucosa. In particular, PRDX1
expression was significantly higher in the lower half of the lamina
propria from the active UC mucosa than that from the normal and
inactive UC mucosae, suggesting its association with inflammation
activity. A previous study observed that monocytes in the blood
release PRDX1 under inflammatory conditions (37). Our results suggest that PRDX1
expression in the epithelium and stromal tissue protects against
oxidative stress in inflammatory foci.
Currently, no reliable biomarker exists for
monitoring disease activity in UC. CRP is often used as a biomarker
of UC activity, but CRP levels are sometimes insufficient to
reflect UC activity. Therefore, biomarkers of UC activity with
sufficient sensitivity and specificity are desired. In recent
studies, serum leucine-rich alpha-2-glycoprotein (LRG)
concentrations correlated with UC activity. Increased LRG
expression was detected in the cytoplasm of epithelial cells in
inflamed tissues. Thus, inflamed colon tissue may be a potential
source of increased serum LRG in patients with UC (38). In addition, other reports showed that
urine levels of prostaglandin E-major urinary metabolite (PGE-MUM)
were significantly correlated with active UC. Furthermore, compared
with CRP levels, PGE-MUM demonstrated increased sensitivity for
reflecting UC activity (39–41). However, these markers are still being
developed. Our results showed that PRDX1 expression correlated with
UC activity, suggesting its potential for monitoring oxidative
stress in the colonic mucosa in association with UC activity.
In addition, we confirmed TRX expression in the UC
regenerative mucosa. TRX is a redox-regulating protein involved in
cellular redox homeostasis and cell survival. TRX is expressed at
relatively high levels in several cancers (42). Our results showed that TRX expression
increased according to the severity of inflammation in epithelial
cells in the crypts of the UC regenerative mucosa. TRX expression
correlated with UC activity, suggesting its potential as a
biomarker for local oxidative stress related to UC activity.
We confirmed PRDX1 and TRX overexpression in
UC-associated neoplastic lesions. PRDX1 expression was
significantly higher in UC-associated neoplastic lesions than in
the normal mucosa, and it increased gradually from LGD to invasive
UCCA. Our results are consistent with an earlier report indicating
that PRDX1-positivity scores were significantly higher in
colorectal cancer than in the normal mucosa (36). TRX expression was also significantly
higher in UC-associated neoplastic lesions than in the normal
mucosa. In a previous study, TRX levels were elevated in
cisplatin-resistant gastric and colon cancer cells (42). Our results showed that both PRDX1 and
TRX proteins were highly expressed in UC-associated neoplastic
lesions, and suggest that the proteins may indicate the presence of
UCCA with oxidative stress. Since surveillance by endoscopic
examination sometimes fails to detect cancer lesions (8), easily measurable biomarkers of UCCA are
desired. Although it is necessary to confirm that PRDX1 and TRX are
released from the UC regenerative mucosa and UCCA cells into the
serum, measuring these proteins in serum samples may help the
clinical detection of malignancy and inflammation activity in the
future.
In this study, two electrophoretic spots of PRDX1
with different PIs were identified using agarose-2DE; the reduced
form of PRDX1 was more highly expressed in active UC biopsy
specimens, and the oxidized form was more highly expressed in
inactive UC and normal biopsy specimens. Moreover, reduced PRDX1
was overexpressed in colon cancer cell lines (UCCA-24, KE-43P)
(data not shown). Thus, PRDX1 expression and oxidative stress are
related in UC inflammatory foci and colon cancer cell lines. PRDX1
expression should be considered not only in terms of
increase/decrease, but also in terms of the kinetics of
reduced/oxidized PRDX1, which can be visualized by agarose-2DE
based on the PIs. As a preliminary study, we exposed UCCA 24 cells
to 200 µM H2O2 for 20 min, which resulted in
PRDX1 upregulation (data not shown). Exposure to
H2O2 enhanced the expression levels of
PRDX1-6 in human umbilical vein endothelial cells (43) and PRDX1 expression in normal human
colon cells (36). PRDX1 and TRX
expression suggests the presence of oxidative stress in the UC
background mucosa, and these two proteins may be involved in UCCA
carcinogenesis. Our study provides insights into carcinogenic
pathways involving chronic inflammation in patients with UC.
In conclusion, PRDX1 and TRX are expressed in the UC
inflamed mucosa and reflect the degree of inflammation. PRDX1 and
TRX overexpression is a unique characteristic of UC-associated
neoplastic lesions, including UCCA, and may reflect oxidative
stress from inflammatory processes in UC. Further studies are
warranted to identify the mechanisms underlying PRDX1 and TRX
functions in UC-associated carcinogenesis with oxidative stress and
to identify novel therapeutic targets for the treatment of
UC-associated neoplastic lesions.
Acknowledgements
The authors would like to thank Dr. Ryo Nagashio for
proteomic analysis support. This study was partially supported by a
project grant for post-graduate students from Kitasato University
Graduate School of Medical Sciences.
References
|
1
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ohshima H, Tazawa H, Sylla BS and Sawa T:
Prevention of human cancer by modulation of chronic inflammatory
processes. Mutat Res. 591:110–122. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Okayasu I: Development of ulcerative
colitis and its associated colorectal neoplasia as a model of the
organ-specific chronic inflammation-carcinoma sequence. Pathol Int.
62:368–380. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wong NA and Harrison DJ: Colorectal
neoplasia in ulcerative colitis-recent advances. Histopathology.
39:221–234. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Okayasu I, Hana K, Yoshida T, Mikami T,
Kanno J and Fujiwara M: Significant increase of colonic mutated
crypts in ulcerative colitis correlatively with duration of
illness. Cancer Res. 62:2236–2238. 2002.PubMed/NCBI
|
|
6
|
Rhodes JM and Campbell BJ: Inflammation
and colorectal cancer: Ibd-associated and sporadic cancer compared.
Trends Mol Med. 8:10–16. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sada M, Igarashi M, Yoshizawa S, Kobayashi
K, Katsumata T, Saigenji K, Otani Y, Okayasu I and Mitomi H: Dye
spraying and magnifying endoscopy for dysplasia and cancer
surveillance in ulcerative colitis. Dis Colon Rectum. 47:1816–1823.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fujii S, Katsumata D and Fujimori T:
Limits of diagnosis and molecular markers for early detection of
ulcerative colitis-associated colorectal neoplasia. Digestion. 77
Suppl 1:S2–S12. 2008. View Article : Google Scholar
|
|
9
|
Vermeire S, Van Assche G and Rutgeerts P:
C-reactive protein as a marker for inflammatory bowel disease.
Inflamm Bowel Dis. 10:661–665. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sachar DB, Smith H, Chan S, Cohen LB,
Lichtiger S and Messer J: Erythrocytic sedimentation rate as a
measure of clinical activity in inflammatory bowel disease. J Clin
Gastroenterol. 8:647–650. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gabay C: Interleukin-6 and chronic
inflammation. Arthritis Res Ther. 8 Suppl 2:S32006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Braegger CP, Nicholls S, Murch SH,
Stephens S and MacDonald TT: Tumour necrosis factor alpha in stool
as a marker of intestinal inflammation. Lancet. 339:89–91. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song M, Wu K, Ogino S, Fuchs CS,
Giovannucci EL and Chan AT: A prospective study of plasma
inflammatory markers and risk of colorectal cancer in men. Br J
Cancer. 108:1891–1898. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schoepfer AM, Beglinger C, Straumann A,
Trummler M, Vavricka SR, Bruegger LE and Seibold F: Fecal
calprotectin correlates more closely with the simple endoscopic
score for crohn's disease (SES-CD) than CRP, blood leukocytes and
the CDAI. Am J Gastroenterol. 105:162–169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gisbert JP and McNicholl AG: Questions and
answers on the role of faecal calprotectin as a biological marker
in inflammatory bowel disease. Dig Liver Dis. 41:56–66. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lehmann FS, Burri E and Beglinger C: The
role and utility of faecal markers in inflammatory bowel disease.
Therap Adv Gastroenterol. 8:23–36. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Judd TA, Day AS, Lemberg DA, Turner D and
Leach ST: Update of fecal markers of inflammation in inflammatory
bowel disease. J Gastroenterol Hepatol. 26:1493–1499. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
D'Haens G, Ferrante M, Vermeire S, Baert
F, Noman M, Moortgat L, Geens P, Iwens D, Aerden I, Van Assche G,
et al: Fecal calprotectin is a surrogate marker for endoscopic
lesions in inflammatory bowel disease. Inflamm Bowel Dis.
18:2218–2224. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
van Rheenen PF, Van de Vijver E and Fidler
V: Faecal calprotectin for screening of patients with suspected
inflammatory bowel disease: Diagnostic meta-analysis. BMJ.
341:c33692010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Okayasu I, Yoshida T, Mikami T, Hana K,
Yokozawa M, Araki K, Mitsuhashi J, Kikuchi M, Adachi E and Sada M:
Mucosal remodeling in long-standing ulcerative colitis with
colorectal neoplasia: Significant alterations of NCAM+ or
alpha-SMA+ subepithelial myofibroblasts and interstitial cells.
Pathol Int. 59:701–711. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Araki K, Mikami T, Yoshida T, Kikuchi M,
Sato Y, Oh-Ishi M, Kodera Y, Maeda T and Okayasu I: High expression
of hsp47 in ulcerative colitis-associated carcinomas: Proteomic
approach. Br J Cancer. 101:492–497. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Matts SG: The value of rectal biopsy in
the diagnosis of ulcerative colitis. Q J Med. 30:393–407.
1961.PubMed/NCBI
|
|
23
|
Nagashio R, Sato Y, Jiang SX, Ryuge S,
Kodera Y, Maeda T and Nakajima T: Detection of tumor-specific
autoantibodies in sera of patients with lung cancer. Lung Cancer.
62:364–373. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nagashio R, Sato Y, Matsumoto T, Kageyama
T, Satoh Y, Ryuge S, Masuda N, Jiang SX and Okayasu I: Significant
high expression of cytokeratins 7, 8, 18, 19 in pulmonary large
cell neuroendocrine carcinomas, compared to small cell lung
carcinomas. Pathol Int. 60:71–77. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yamashita K, Yasuda S, Kuba T, Otani Y,
Fujiwara M and Okayasu I: Unique characteristics of rectal
carcinoma cell lines derived from invasive carcinomas in ulcerative
colitis patients. Cancer Sci. 95:211–217. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tokuyama W, Mikami T, Fujiwara M, Matsui T
and Okayasu I: Midkine expression in colorectal tumors: Correlation
with ki-67 labeling in sporadic, but not ulcerative
colitis-associated ones. Pathol Int. 57:260–267. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rhee SG and Woo HA: Multiple functions of
peroxiredoxins: Peroxidases, sensors and regulators of the
intracellular messenger H2O2 and protein chaperones. Antioxid Redox
Signal. 15:781–794. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Karihtala P, Mantyniemi A, Kang SW,
Kinnula VL and Soini Y: Peroxiredoxins in breast carcinoma. Clin
Cancer Res. 9:3418–3424. 2003.PubMed/NCBI
|
|
29
|
Poynton RA and Hampton MB: Peroxiredoxins
as biomarkers of oxidative stress. Biochim Biophys Acta.
1840:906–912. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang B, Wang Y and Su Y: Peroxiredoxins,
a novel target in cancer radiotherapy. Cancer Lett. 286:154–160.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cha MK, Suh KH and Kim IH: Overexpression
of peroxiredoxin I and thioredoxin1 in human breast carcinoma. J
Exp Clin Cancer Res. 28:932009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou J, Shen W, He X, Qian J, Liu S and Yu
G: Overexpression of prdx1 in hilar cholangiocarcinoma: A predictor
for recurrence and prognosis. Int J Clin Exp Pathol. 8:9863–9874.
2015.PubMed/NCBI
|
|
33
|
Hoshino I, Matsubara H, Akutsu Y,
Nishimori T, Yoneyama Y, Murakami K, Sakata H, Matsushita K and
Ochiai T: Tumor suppressor prdx1 is a prognostic factor in
esophageal squamous cell carcinoma patients. Oncol Rep. 18:867–871.
2007.PubMed/NCBI
|
|
34
|
Ambrosone CB: Oxidants and antioxidants in
breast cancer. Antioxid Redox Signal. 2:903–917. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ding C, Fan X and Wu G: Peroxiredoxin 1-an
antioxidant enzyme in cancer. J Cell Mol Med. 21:193–202. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chu G, Li J, Zhao Y, Liu N, Zhu X, Liu Q,
Wei D and Gao C: Identification and verification of prdx1 as an
inflammation marker for colorectal cancer progression. Am J Transl
Res. 8:842–859. 2016.PubMed/NCBI
|
|
37
|
Liu CH, Kuo SW, Hsu LM, Huang SC, Wang CH,
Tsai PR, Chen YS, Jou TS and Ko WJ: Peroxiredoxin 1 induces
inflammatory cytokine response and predicts outcome of cardiogenic
shock patients necessitating extracorporeal membrane oxygenation:
An observational cohort study and translational approach. J Transl
Med. 14:1142016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Serada S, Fujimoto M, Terabe F, Iijima H,
Shinzaki S, Matsuzaki S, Ohkawara T, Nezu R, Nakajima S, Kobayashi
T, et al: Serum leucine-rich alpha-2 glycoprotein is a disease
activity biomarker in ulcerative colitis. Inflamm Bowel Dis.
18:2169–2179. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Arai Y, Arihiro S, Matsuura T, Kato T,
Matsuoka M, Saruta M, Mitsunaga M, Matsuura M, Fujiwara M, Okayasu
I, et al: Prostaglandin e-major urinary metabolite as a reliable
surrogate marker for mucosal inflammation in ulcerative colitis.
Inflamm Bowel Dis. 20:1208–1216. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Arai Y, Matsuura T, Matsuura M, Fujiwara
M, Okayasu I, Ito S and Arihiro S: Prostaglandin e-major urinary
metabolite as a biomarker for inflammation in ulcerative colitis:
Prostaglandins revisited. Digestion. 93:32–39. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hagiwara SI, Okayasu I, Fujiwara M,
Matsuura M, Ohnishi H, Ito S, Kishimoto H, Nambu R and Kagimoto S:
Prostaglandin e-major urinary metabolite as a biomarker for
pediatric ulcerative colitis activity. J Pediatr Gastroenterol
Nutr. 64:955–961. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Marks PA: Thioredoxin in cancer-role of
histone deacetylase inhibitors. Semin Cancer Biol. 16:436–443.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mitsumoto A, Takanezawa Y, Okawa K,
Iwamatsu A and Nakagawa Y: Variants of peroxiredoxins expression in
response to hydroperoxide stress. Free Radic Biol Med. 30:625–635.
2001. View Article : Google Scholar : PubMed/NCBI
|