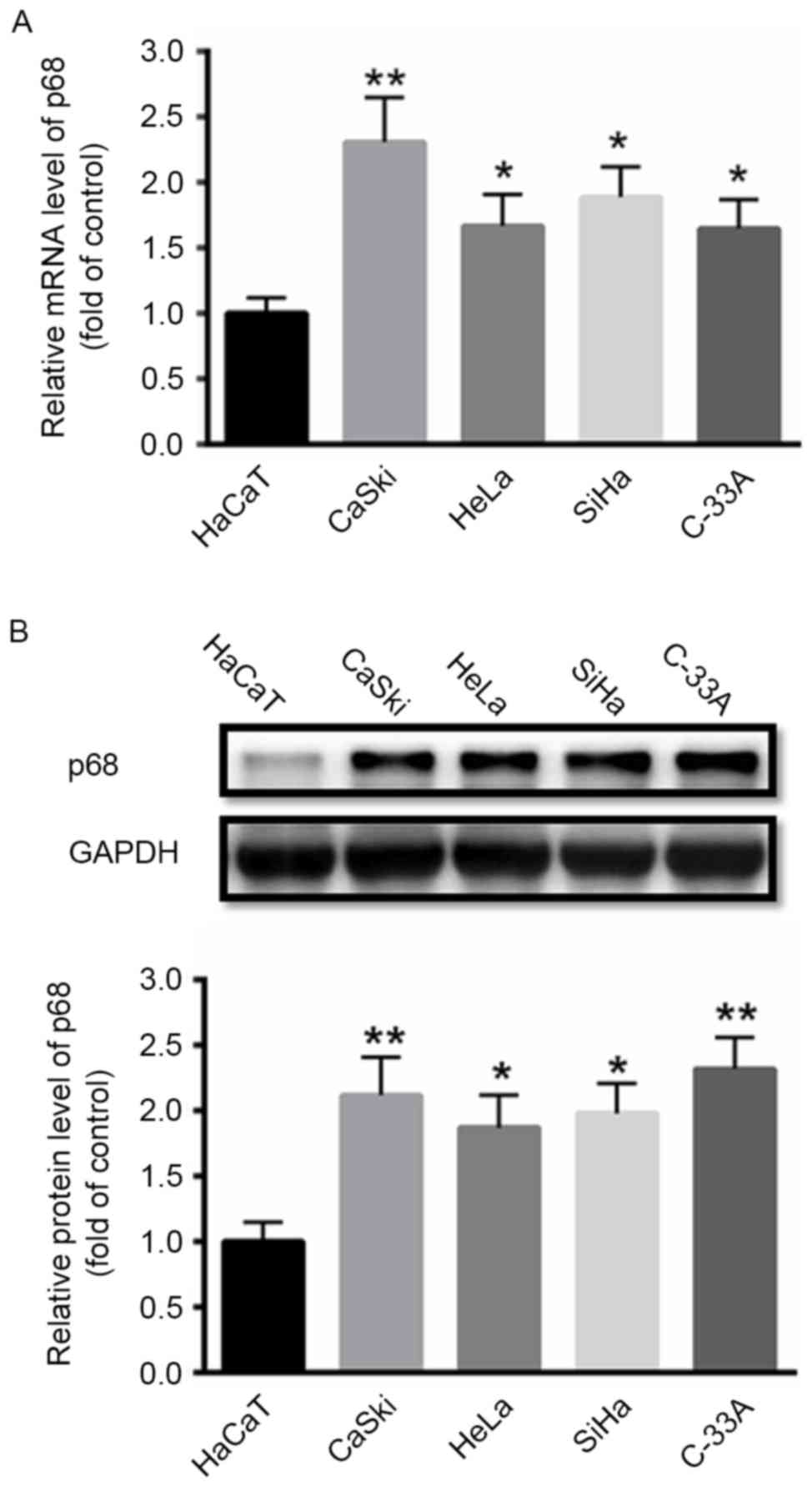
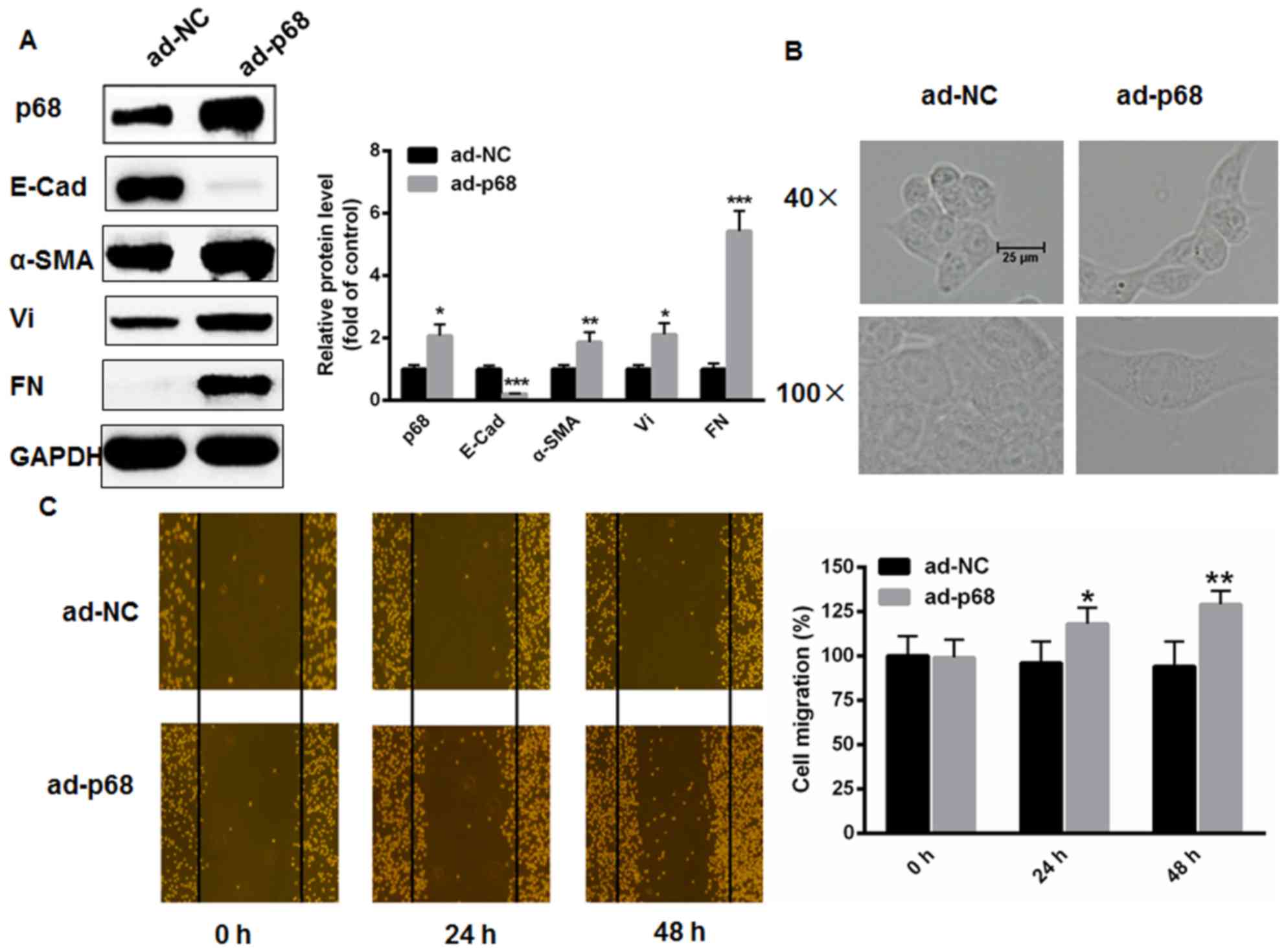
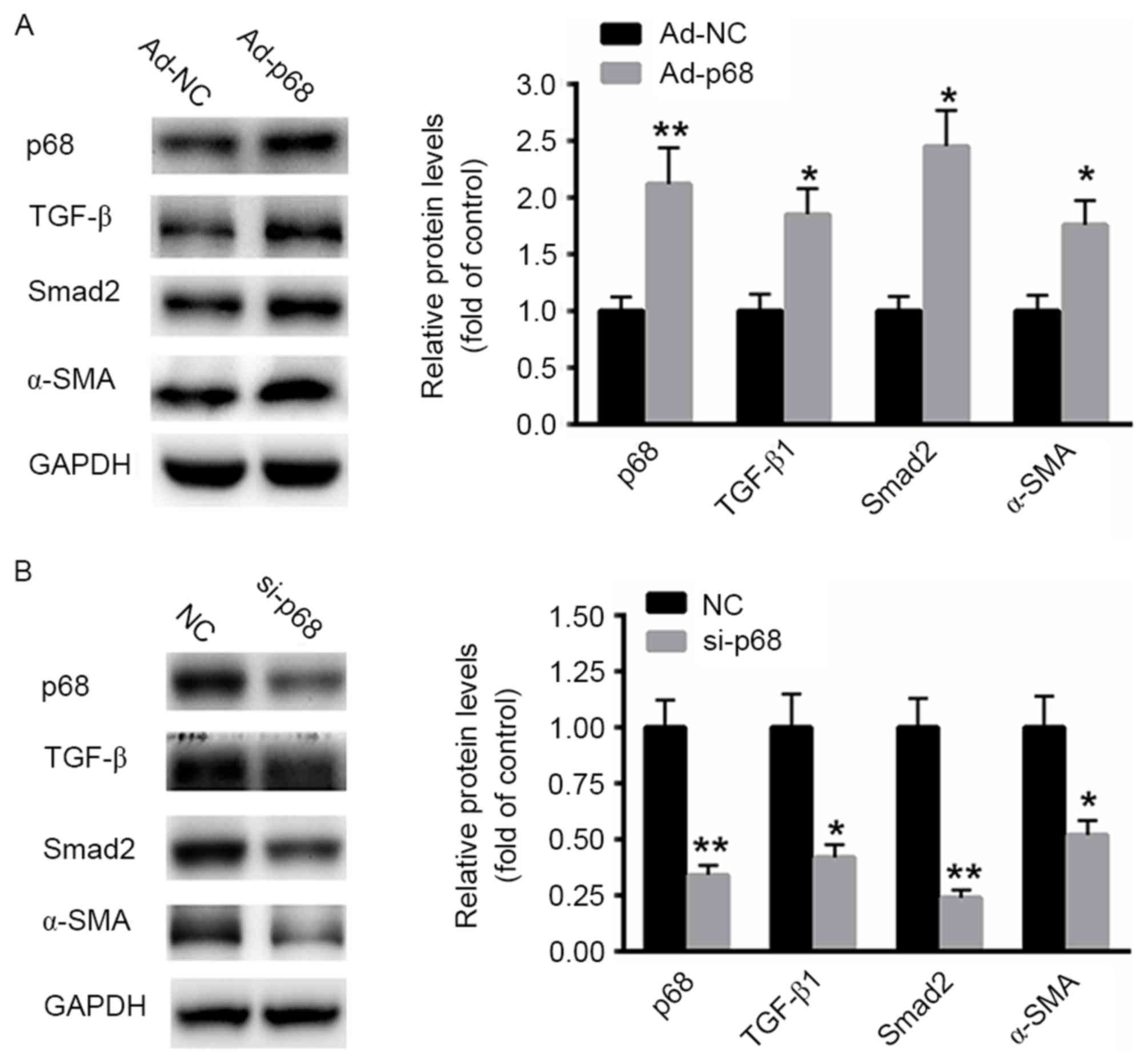
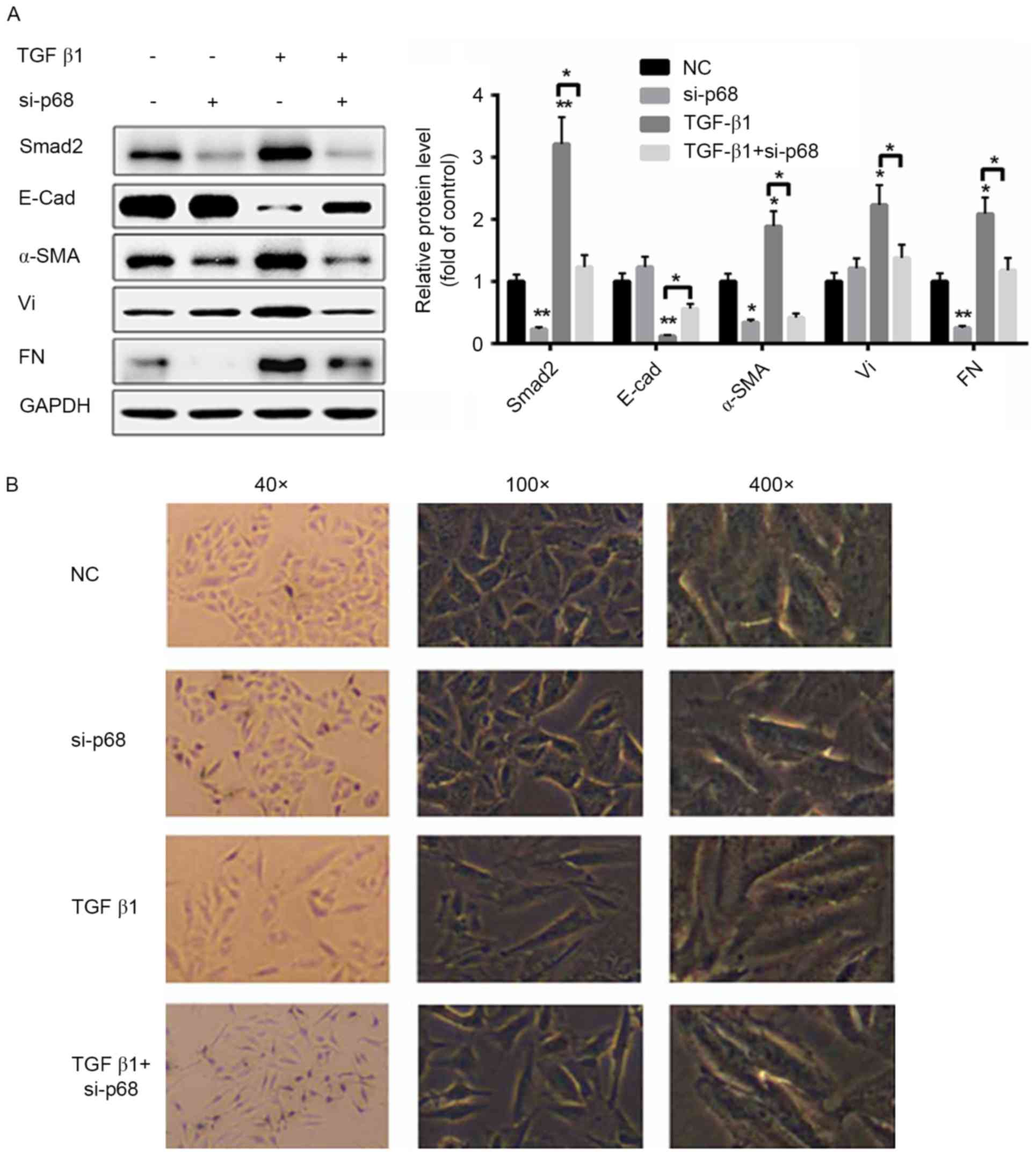
|
1
|
Kim M, Kim YS, Kim H, Kang MY, Park J, Lee
DH, Roh GS, Kim HJ, Kang SS, Cho GJ, et al: O-linked
N-acetylglucosamine transferase promotes cervical cancer
tumorigenesis through human papillomaviruses E6 and E7 oncogenes.
Oncotarget. 7:44596–44607. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN, 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Caramel J, Papadogeorgakis E, Hill L,
Browne GJ, Richard G, Wierinckx A, Saldanha G, Osborne J,
Hutchinson P, Tse G, et al: A switch in the expression of embryonic
EMT-inducers drives the development of malignant melanoma. Cancer
Cell. 24:466–480. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fenouille N, Tichet M, Dufies M, Pottier
A, Mogha A, Soo JK, Rocchi S, Mallavialle A, Galibert MD, Khammari
A, et al: The epithelial-mesenchymal transition (EMT) regulatory
factor SLUG (SNAI2) is a downstream target of SPARC and AKT in
promoting melanoma cell invasion. PLoS One. 7:e403782012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang GM, Xie WY, Wang HS, Du J, Wu BP, Xu
W, Liu HF, Xiao P, Liu ZG, Li HY, et al: Curcumin combined with
FAPαc vaccine elicits effective antitumor response by targeting
indolamine-2,3-dioxygenase and inhibiting EMT induced by TNF-α in
melanoma. Oncotarget. 6:25932–25942. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jung HY, Fattet L and Yang J: Molecular
pathways: Linking tumor microenvironment to epithelial-mesenchymal
transition in metastasis. Clin Cancer Res. 21:962–968. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang P, Sun Y and Ma L: ZEB1: At the
crossroads of epithelial-mesenchymal transition, metastasis and
therapy resistance. Cell Cycle. 14:481–487. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Laurenzana A, Biagioni A, Bianchini F,
Peppicelli S, Chillà A, Margheri F, Luciani C, Pimpinelli N, Del
Rosso M, Calorini L and Fibbi G: Inhibition of uPAR-TGFβ crosstalk
blocks MSC-dependent EMT in melanoma cells. J Mol Med (Berl).
93:783–794. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin K, Baritaki S, Militello L, Malaponte
G, Bevelacqua Y and Bonavida B: The role of B-RAF mutations in
melanoma and the induction of EMT via DYsregulation of the
NF-κB/Snail/RKIP/PTEN circuit. Genes Cancer. 1:409–420. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schlegel NC, von Planta A, Widmer DS,
Dummer R and Christofori G: PI3K signalling is required for a
TGFβ-induced epithelial-mesenchymal-like transition (EMT-like) in
human melanoma cells. Exp Dermatol. 24:22–28. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tulchinsky E, Pringle JH, Caramel J and
Ansieau S: Plasticity of melanoma and EMT-TF reprogramming.
Oncotarget. 5:1–2. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lane DP and Hoeffler WK: SV40 large T
shares an antigenic determinant with a cellular protein of
molecular weight 68,000. Nature. 288:167–170. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fukuda T, Yamagata K, Fujiyama S,
Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H,
Nakamura T, et al: DEAD-box RNA helicase subunits of the drosha
complex are required for processing of rRNA and a subset of
microRNAs. Nat Cell Biol. 9:604–611. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Abdelhaleem M: RNA helicases: Regulators
of differentiation. Clin Biochem. 38:499–503. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fuller-Pace FV: DExD/H box RNA helicases:
Multifunctional proteins with important roles in transcriptional
regulation. Nucleic Acids Res. 34:4206–4215. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Metivier R, Penot G, Hubner MR, Reid G,
Brand H, Kos M and Gannon F: Estrogen receptor-alpha directs
ordered, cyclical, and combinatorial recruitment of cofactors on a
natural target promoter. Cell. 115:751–763. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Clark EL, Coulson A, Dalgliesh C, Rajan P,
Nicol SM, Fleming S, Heer R, Gaughan L, Leung HY, Elliott DJ, et
al: The RNA helicase p68 is a novel androgen receptor coactivator
involved in splicing and is overexpressed in prostate cancer.
Cancer Res. 68:7938–7946. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shin S, Rossow KL, Grande JP and Janknecht
R: Involvement of RNA helicases p68 and p72 in colon cancer. Cancer
Res. 67:7572–7578. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang L, Lin C and Liu ZR: P68 RNA helicase
mediates PDGF-induced epithelial mesenchymal transition by
displacing Axin from beta-catenin. Cell. 127:139–155. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang L, Lin C, Zhao S, Wang H and Liu ZR:
Phosphorylation of p68 RNA helicase plays a role in
platelet-derived growth factor-induced cell proliferation by
up-regulating cyclin D1 and c-Myc expression. J Biol Chem.
282:16811–16819. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo J, Li M, Meng X, Sui J, Dou L, Tang W,
Huang X, Man Y, Wang S, Li J, et al: MiR-291b-3p induces apoptosis
in liver cell line NCTC1469 by reducing the level of RNA-binding
protein HuR. Cell Physiol Biochem. 33:810–822. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gupta GP and Massagué J: Cancer
metastasis: Building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fuller-Pace FV: RNA helicases: Modulators
of RNA structure. Trends Cell Biol. 4:271–274. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schmid SR and Linder P: D-E-A-D protein
family of putative RNA helicases. Mol Microbiol. 6:283–291. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Song C, Hotz-Wagenblatt A, Voit R and
Grummt I: SIRT7 and the DEAD-box helicase DDX21 cooperate to
resolve genomic R loops and safeguard genome stability. Genes Dev.
2017. View Article : Google Scholar
|
|
27
|
Lumb JH, Li Q, Popov LM, Ding S, Keith MT,
Merrill BD, Greenberg HB, Li JB and Carette JE: DDX6 represses
aberrant activation of interferon-stimulated genes. Cell Rep.
20:819–831. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mazurek A, Luo W, Krasnitz A, Hicks J,
Powers RS and Stillman B: DDX5 regulates DNA replication and is
required for cell proliferation in a subset of breast cancer cells.
Cancer Discov. 2:812–825. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Westphal P, Mauch C, Florin A, Czerwitzki
J, Olligschläger N, Wodtke C, Schüle R, Büttner R and Friedrichs N:
Enhanced FHL2 and TGF-β1 expression is associated with invasive
growth and poor survival in malignant melanomas. Am J Clin Pathol.
143:248–256. 2015. View Article : Google Scholar : PubMed/NCBI
|