Introduction
The association between inflammation and the
development of cancer has been suggested for numerous years
(1,2).
Specifically, in breast cancer inflammation is increasingly
recognized as an important component of tumorigenesis. Previous
studies have reported that numerous inflammatory mediators
influence breast cancer development and progression (3–7).
There is increasing evidence that cancer stem cells
(CSCs) mediate tumor growth and metastasis (8). CSCs possess two main properties, the
ability to self-renew and the ability to differentiate into
heterogeneous lineages of cancer cells that comprise the tumor.
Breast tumors cells that exhibit the properties of CSCs have been
termed breast CSCs (BCSCs) (9,10).
Previously, it has been suggested that inflammation may regulate
BCSCs, and certain immune mediators have been reported to influence
BCSC biology (11). There is an
investigation into immunotherapies targeting CSCs and an initial
report has demonstrated the potential of immunotherapy as a cancer
treatment (12); however, the
mechanisms underlying these approaches are not yet fully
characterized. The majority of previous studies were pre-clinical,
and the role of inflammation and CSCs in patients with breast
cancer was not well defined. In the present study, the association
between inflammation and the BCSC phenotype was evaluated in human
breast cancer tissue. In addition, the association between BCSCs
and inflammation in the progression of breast cancer was
investigated.
Materials and methods
Patients and tissue microarrays
(TMAs)
A total of 47 consecutive patients with primary
breast cancer who had undergone surgery between May 2008 and
November 2011 at Daegu Catholic University Hospital (Daegu, Korea)
were included in the present study. The inclusion criteria were as
follows: i) Patient had primary breast cancer; ii) patient provided
informed consent; iii) patient had undergone surgery, including
breast conserving surgery or mastectomy; iv) patient had a tissue
sample available following surgery. All patients were female; the
mean age of the patients was 55.77±13.47 years (range, 34–90
years).
All data was retrospectively analyzed. All tissue
specimens had previously been formalin fixed, paraffin embedded,
stained with hematoxylin and eosin, and reviewed by an experienced
pathologist. The clinical information of the patients and their
tumor characteristics, including tumor size, nodal status,
histological grade, lymphovascular invasion status and other
prognostic factors, were evaluated based on medical records, and
pathological reports. Breast cancer staging was assessed according
to the seventh edition of the American Joint Committee on Cancer
staging manual for breast cancer (13). Histologic grade was assessed using the
Nottingham grading system (14).
Ethical approval for the study was obtained from the Institutional
Review Board of Daegu Catholic University Hospital. Written
informed consent was obtained from all patients.
TMAs were constructed using representative paraffin
blocks of 47 cases of invasive breast carcinoma and 10 normal
breast tissue samples obtained from the same patients, following
the method described in our previous study (15).
Immunohistochemical staining
Immunohistochemical staining was performed on TMA
sections, including cancer and normal tissue, using the Bond
Polymer Intense Detection system (Leica Microsystems, Inc., Buffalo
Grove, IL, USA) according to the manufacturer's protocol with minor
modifications. The TMA blocks were cut into 5-µm-thick sections and
deparaffinized with Bond Dewax solution (Leica Microsystems, Inc.).
An antigen retrieval procedure was performed using Bond ER Solution
(Leica Microsystems, Inc.) for 30 min at 100°C.
Endogenous peroxidase activity was quenched with
hydrogen peroxide for 5 min at 25°C. Sections were then incubated
for 15 min at room temperature with primary monoclonal antibodies
directed against the following proteins: cluster of differentiation
(CD)24 (dilution, 1:20; cat. no. SC-7034; clone C-20; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), CD44 (dilution, 1:1,000;
cat. no. NBP1-47386; clone, 8E2F3; Novus Biologicals, LLC,
Littleton, CO, USA), CD4 (ready-to-use dilution; cat. no. PA0368;
clone 4B12; Leica Biosystems, Inc., Wetzlar, Germany), CD8
(dilution 1:200; cat. no. M7103; clone C8/144B), CD68 (dilution,
1:200; cat no. M0876; clone, PG-M1), epidermal growth factor
receptor (EGFR; dilution, 1:100; cat. no. M7239; clone, EGFR.25),
apoptosis regulator Bcl-2 (dilution, 1:4; cat. no. IR614; clone,
124), human epidermal growth factor receptor 2 (HER2; dilution,
1:250; cat. no. A048529-1; clone, A0485; all from Dako; Agilent
Technologies, Inc.), estrogen receptor (ER; dilution, 1:100; cat.
no. NCL-L-ER-6F11; clone, 6F11), progesterone receptor (PR;
dilution, 1:100; cat. no. NCL-L-PGR-312; clone, 16; both from
Novocastra; Leica Biosystems, Inc.), proliferation marker protein
Ki-67 (dilution, 1:200; cat. no. 275R-16; clone, MM1-L;
Sigma-Aldrich; Merck KGaA) and tumor antigen p53 (dilution, 1:200;
cat. no. 18-0129; clone, BP53.12; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA).
The samples were then treated with a biotin-free
polymeric horseradish peroxidase-linker antibody conjugate system
(Bond Polymer Refine Detection; ready-to-use dilution; cat. no.
DS9800; Leica Biosystems, Inc.). Staining was performed in a
Bond-Max Automatic Slide Stainer (Leica Microsystems, Inc.). A BX50
light microscope (Olympus Corporation, Tokyo, Japan) was used to
visualize staining at ×400 magnification; the stained cells were
manually counted.
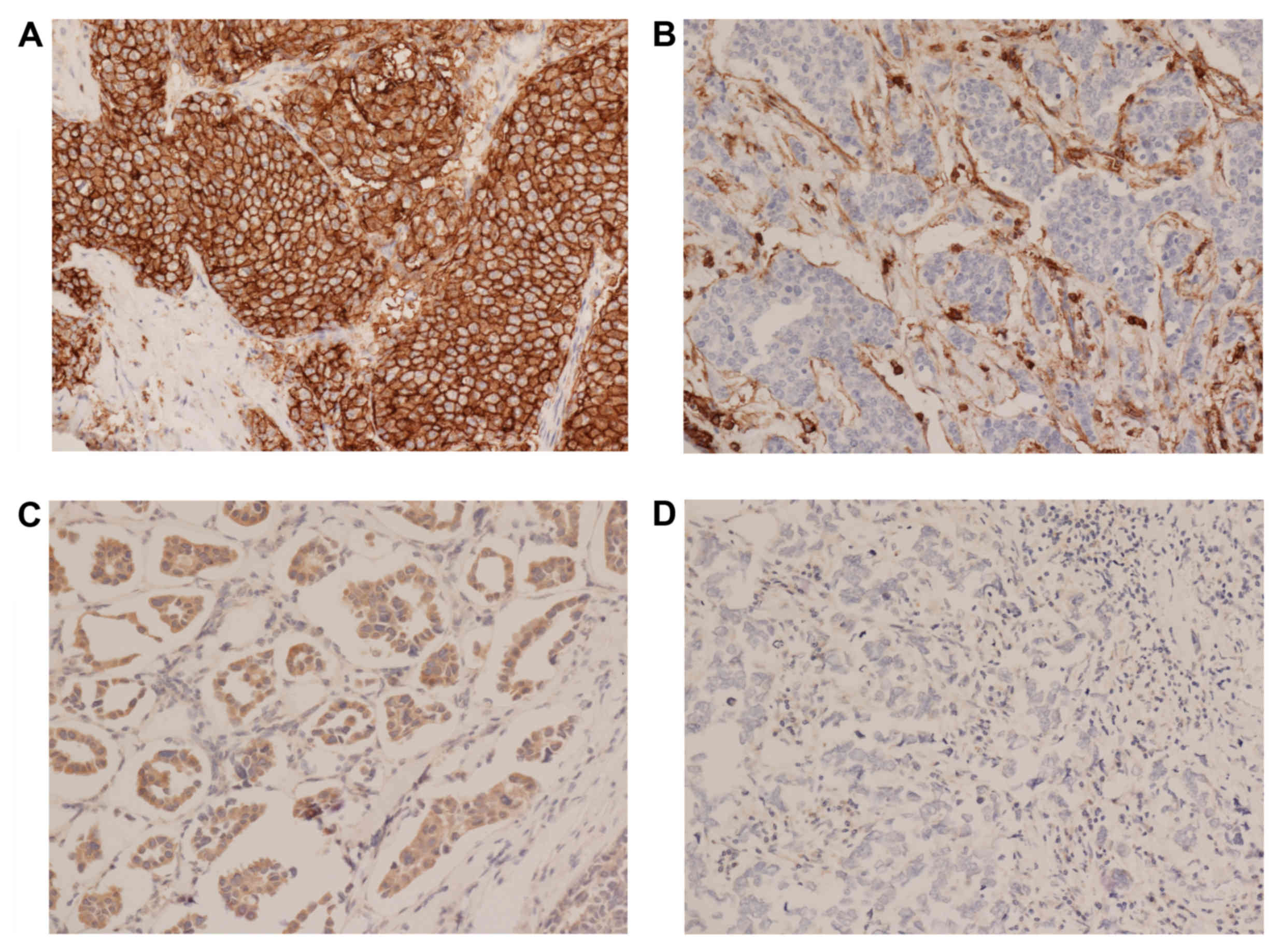
Expression levels of CD24 and CD44 were graded on
staining intensity and the proportion of positively stained tumor
cells. The levels of immunopositivity were semiquantitatively
scored as follows: 0, No staining; 1+, minimal staining
intensity, <10% of cells positively stained; 2+,
moderate staining intensity, 10–50% of cells positively stained and
3+, marked, staining intensity, >50% of cells
positively stained. Scores of 0 and 1 were designated as negative,
and 2 and 3 as positive. Examples of this staining are illustrated
in Fig. 1. BCSCs were defined as
CD44+/CD24− tumor cells. For ER and PR,
nuclear staining in ≥1% of tumor cells was considered positive.
Cytoplasmic and membranous staining of any intensity in ≥5% of the
tumor cells was considered as positive for Bcl-2. Membranous
staining for HER2 with strong complete staining in 30% of the tumor
cells was regarded as HER2 overexpression. p53 staining was scored
positive if ≥5% of the cells were stained with a strong intensity.
The Ki-67 labeling index was expressed as a percentage and was
graded as high if the number of positively stained cells was
≥14%.
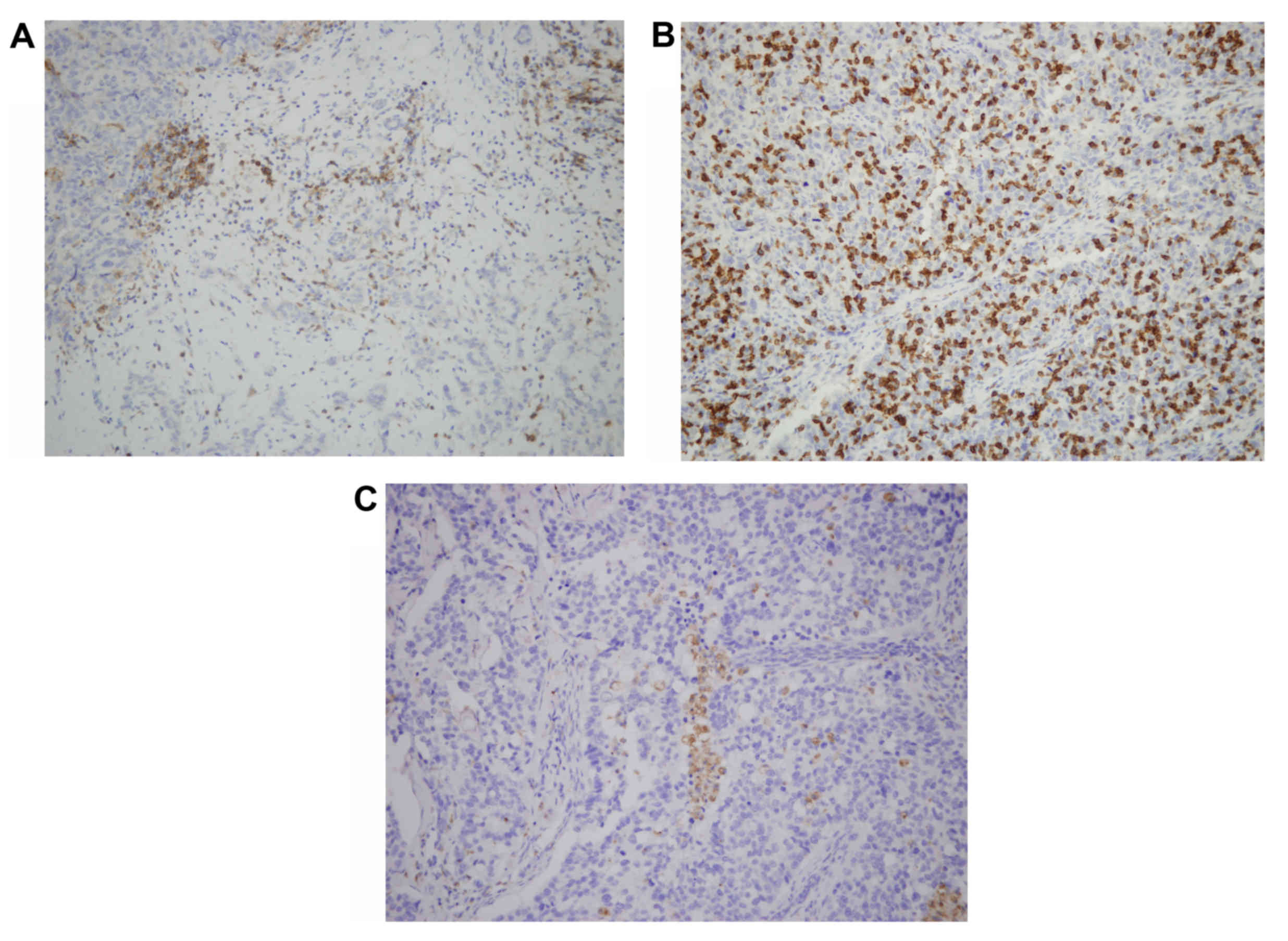
The CD4, CD8 and CD68 immunostained TMA sections
were evaluated under a microscope and the number of CD4+
and CD8+ T cells and CD68+ macrophages were
counted in the stroma and cancer cell nests. Examples of this
staining are illustrated in Fig. 2.
Intratumoral (in the tumor cell nest) or peritumoral (in the stroma
around the tumor) lymphocyte infiltration was semiquantitatively
graded as follows: 0, No lymphocyte infiltration; 1, mild scattered
lymphocyte infiltration in either stroma or tumor cell nest; 2,
moderate lymphocyte infiltration with some lymph follicle
formation; 3, dense and widespread lymphocyte infiltration.
Reverse transcription polymerase chain
reaction (RT-PCR)
The levels of inflammatory modulators and cytokines,
including tumor necrosis factor (TNF)-α, interleukin (IL)-2, −4 and
−6, interferon (IFN)-γ and nuclear factor (NF)-κB p50 were assessed
by the levels of mRNA transcripts in frozen tissue using RT-PCR.
Total RNA was extracted from frozen breast cancer tissues using
Trizol reagent (cat. no. A33250; Invitrogen; Thermo Fisher
Scientific, Inc.). Subsequent to lysing and homogenizing samples in
the Trizol reagent, the samples were incubated for 5 min at room
temperature. Chloroform was added to the samples, and the samples
were agitated for 15 sec, then incubated for 2–3 min at room
temperature. Following centrifugation for 5 min at 12,000–16,000 ×
g at 4°C, the RNA in the samples was precipitated by adding
isopropanol. The samples were washed with in 75% ethanol then the
RNA was dissolved with RNase-free water. The RNA was quantified by
measuring absorbance at 260 and 280 nm.
To determine the expression of ALCAM, inflammatory
modulators and cytokines, reverse transcription of the total RNA
was performed. First-strand complementary (c)DNA was generated
using a commercial kit (Superscript II RNase H-reverse
transcriptase, cat no. 18064071; Invitrogen; Thermo Fisher
Scientific, Inc.) used according to the manufacturer's protocol.
For the PCR of ALCAM, TNF-α, IL-4, IFN-γ and NF-κB p50, the
following primers were used: ALCAM, forward,
5′-CAAGACAACCAAGGCTGACA-3′; reverse, 5′-CGCAGACATAGTTTCCAGCA-3′;
TNF-α, forwards, 5′-CCCTCAACCTCTTCTGGCTC-3′; reverse,
5′-AGGCAGCTCCTACATTGGGT−3′; IL-2, forwards,
5′-GCAACTCCTGTCTTGCATTG-3′; reverse, 5′-TGCTTTGACAAAAGGTAATCCA-3′;
IL-4, forwards, 5′-ACTGCTTCCCCCTCTGTTCT-3′; reverse,
5′-TGATCGTCTTTAGCCTTTCCA-3′; IL-6, forwards,
5′-TACCCCCAGGAGAAGATTCC-3′; reverse, 5′-AAAGCTGCGCAGAATGAGAT-3′;
interferon-γ, forwards, 5′-TTGGCTTTTCAGCTCTGCAT-3′; reverse,
5′-CTGTTTTAGCTGCTGGCGAC-3′; NF-kB p50, forwards,
5′-CACCTAGCTGCCAAAGAAGG-3′; reverse, 5′-TCAGCCAGCTGTTTCATGTC-3′.
β-actin was used as a reference gene and the primer was as follows:
Forwards, 5′-AGGGTGTGATGTGGGTATGG-3′; reverse,
5′-CAGGATCTTCATGAGGTAGTC-3′.
PCR was performed with 1 µl of cDNA and 0.4 U Taq
polymerase (cat. no. #18038042; Thermo Fisher Scientific, Inc.).
The thermocycler settings were as follows: An initial temperature
of 94°C for 2 min, then 35 cycles of 94°C for 30 sec, 65°C for 30
sec and 72°C for 1 min. PCR products were analyzed by agarose gel
electrophoresis and visualized with ethidium bromide staining.
Statistical analysis
Statistical analyses were performed using SPSS
software (version 15.0; SPSS, Inc., Chicago, IL, USA). A one-sample
Kolmogorov-Smirnov test was used to evaluate the distribution of
continuous parameters. The association between BCSC phenotype and
the number of inflammatory cells was assessed using a Student's
t-test for CD8+ T cells and CD68+ macrophages
and a non-parametric Mann-Whitney U test for CD4+ T
cells. The association between other inflammatory modulators and
the BCSC phenotype was assessed using a χ2 test for
intratumoral and peritumoral inflammation, and Fisher's exact test
for TNF-α, IL-4 and NF-kB p50 expression status. The association
between the BCSC phenotype and the clinicopathological
characteristics of the patients was analyzed using the Chi-square
test for categorical data, including menopausal state, T stage,
node metastasis, histologic grade, lymphovascular invasion, ER, PR,
Bcl-2, p53 and EGFR expression status, HER2 overexpression status,
Ki-67 index and molecular subtype. All tests were two-tailed.
P<0.05 was considered to indicate a statistically significant
difference.
Results
The clinicopathological characteristics of the
patients included in the present study are illustrated in Table I. The mean age of the patients with
breast cancer was 55.77±13.47 years (range, 34–90 years). All cases
were categorized into four groups according to the
immunohistochemical results for CD44 and CD24 (Table II). Of the 47 patients, 10 (21.3%)
exhibited the BCSC phenotype (CD44+/CD24−).
CD44 positivity was significantly higher in postmenopausal women
compared with in premenopausal women (P=0.004; data not shown).
Intratumoral inflammation was significantly more frequent in the
CD44-negative groups (P=0.018) compared with CD44-positive groups
(data not shown).
 | Table I.Clinicopathological characteristics
of patients with breast cancer. |
Table I.
Clinicopathological characteristics
of patients with breast cancer.
| Clinicopathological
characteristic | Value |
|---|
| Age, years [mean ±
standard deviation (range)] | 55.77±13.47
(34–90) |
| Menopausal status,
n (%) |
|
|
Premenopausal | 17 (38.6) |
|
Postmenopausal | 27 (61.4) |
| Tumor size, cm
[mean ± standard deviation (range)] | 1.97±1.04
(0.10–4.50) |
| Histologic grade, n
(%) |
|
| I | 8 (17.0) |
| II | 12 (25.5) |
|
III | 27 (57.5) |
| Nodal involvement,
n (%) |
|
|
Negative | 29 (61.7) |
|
Positive | 18 (38.3) |
| Distant metastasis,
n (%) |
|
|
Negative | 45 (95.7) |
|
Positive | 2 (4.3) |
| Tumor stage, n
(%) |
|
| I | 18 (40.0) |
|
IIA | 15 (33.3) |
|
IIB | 9 (20.0) |
|
IIIA | 2 (4.5) |
|
IIIB | 0 (0.0) |
|
IIIC | 1 (2.2) |
| IV | 0 (0.0) |
| Molecular subtype,
n (%) |
|
| Luminal
A | 9 (22.0) |
| Luminal
B | 23 (56.1) |
|
HER2 | 6 (14.6) |
|
Basal-like | 3 (7.3) |
| Lymphovascular
invasion, n (%) |
|
|
Negative | 27 (57.4) |
|
Positive | 20 (42.6) |
| ER expression
status, n (%) |
|
|
Negative | 15 (31.9) |
|
Positive | 32 (68.1) |
| PR expression
status, n (%) |
|
|
Negative | 10 (21.3) |
|
Positive | 37 (78.7) |
| HER2 overexpression
status, n (%) |
|
|
Negative | 24 (57.1) |
|
Positive | 18 (42.9) |
| Ki-67 index, n
(%) |
|
|
<14% | 2 (4.3) |
|
≥14% | 44 (95.7) |
 | Table II.Immunohistochemical staining results
for CD44 and CD24. |
Table II.
Immunohistochemical staining results
for CD44 and CD24.
| Tumor cell
phenotype | No. of patients
(%) |
|---|
|
CD44+/CD24− | 10
(21.3) |
|
CD44+/CD24+ | 9 (19.1) |
|
CD44−/CD24+ | 22
(46.8) |
|
CD44−/CD24− | 6 (12.8) |
A CD44+/CD24− phenotype was
significantly inversely associated with lymph node metastasis
(P=0.038; Table III). The
CD44+/CD24− phenotype was also significantly
associated with the molecular subtype of breast cancer (P=0.042),
being particularly more abundant in the basal-like subtype
(Table III). In addition, the
presence of CD44+/CD24− tumor cells was
associated with intratumoral inflammation (P=0.032; Table III) and tumor-infiltrating
CD4+ T cell counts (P=0.003; Table IV).
 | Table III.Clinicopathological characteristics
associated with a CD44+/CD24− phenotype in
invasive breast cancer tissue samples. |
Table III.
Clinicopathological characteristics
associated with a CD44+/CD24− phenotype in
invasive breast cancer tissue samples.
| Clinicopathological
characteristic |
CD44+/CD24− positive
patients (%) | P-value |
|---|
| Menopausal
state |
| 0.057 |
|
Pre-menopausal | 5.9 |
|
|
Post-menopausal | 29.6 |
|
| T stage |
| 0.366 |
|
≤T1 | 25.9 |
|
|
≥T2 | 15.0 |
|
| Node
metastasis |
| 0.038 |
|
Negative | 31.0 |
|
|
Positive | 5.6 |
|
| Histologic
grade |
| 0.092 |
| 1 | 50.0 |
|
| 2 | 16.7 |
|
| 3 | 14.8 |
|
| Lymphovascular
invasion |
| 0.366 |
|
Negative | 25.9 |
|
|
Positive | 15.0 |
|
| ER expression
status |
| 0.536 |
|
Negative | 26.7 |
|
|
Positive | 18.8 |
|
| PR expression
status |
| 0.103 |
|
Negative | 40.0 |
|
|
Positive | 16.2 |
|
| HER2 overexpression
status |
| 0.347 |
|
Negative | 29.2 |
|
|
Positive | 16.7 |
|
| Bcl-2 expression
status |
| 0.778 |
|
Negative | 25.0 |
|
|
Positive | 20.5 |
|
| p53 expression
status |
| 0.609 |
|
Negative | 28.6 |
|
|
Positive | 20.0 |
|
| Ki-67 index |
| 0.322 |
|
<10% | 50.0 |
|
|
≥10% | 20.5 |
|
| EGFR expression
status |
| 0.579 |
|
Negative | 19.4 |
|
|
Positive | 27.3 |
|
| Molecular
subtype |
| 0.042 |
| Luminal
A | 44.4 |
|
| Luminal
B | 8.7 |
|
|
HER2 | 33.3 |
|
|
Basal-like | 66.7 |
|
| Intratumoral
inflammation |
| 0.032 |
|
Negative | 50.0 |
|
|
Positive | 14.3 |
|
| Peritumoral
inflammation |
| 0.156 |
|
Negative | 50.0 |
|
|
Positive | 18.8 |
|
 | Table IV.Association between a
CD44+/CD24− phenotype and inflammatory
markers in invasive breast cancer tissue samples. |
Table IV.
Association between a
CD44+/CD24− phenotype and inflammatory
markers in invasive breast cancer tissue samples.
|
|
CD44+/CD24− tumor
cell phenotype |
|---|
|
|
|
|---|
| Inflammatory
marker | Positive | Negative | P-value |
|---|
| TNF-α expression
status, n |
|
|
|
|
Negative | 2 | 13 | 0.362 |
|
Positive | 8 | 24 |
|
| IL-4 expression
status, n |
|
|
|
|
Negative | 4 | 18 | 0.627 |
|
Positive | 6 | 19 |
|
| NF-κB p50
expression status, n |
|
|
|
|
Negative | 0 | 2 | 0.452 |
|
Positive | 10 | 35 |
|
| CD4+ T
cell count, mean | 9.2 | 31.7 | 0.003 |
| CD8+ T
cell count, mean | 64.0 | 120.3 | 0.110 |
| CD68+ macrophage
count, mean | 25.6 | 31.9 | 0.505 |
Analysis of the clinicopathological significance of
inflammatory mediators and inflammatory cells demonstrated that
tumor-infiltrating CD8+ T cells were significantly
increased in patients with basal-like subtype of breast cancer
(P=0.037) compared with other molecular subtypes (data not
shown).
Discussion
There is increasing evidence that inflammation and
CSCs are associated with carcinogenesis in numerous tumor types
(11,16–19).
Recent studies have suggested an association between inflammation
within the tumor microenvironment and CSCs (17,19);
however, the effect of inflammation on CSCs has yet to be fully
determined. Blaylock (19) reported
that inflammation is essential to cancer induction through its
mutagenic effects on stem cell DNA. Shigdar et al (17) demonstrated that inflammatory response
and stimuli from immune cells, including cytokines, cause cancer
cells to dedifferentiate into CSCs through several signaling
pathways, including the NF-κB signaling pathway. In breast cancer,
several studies have reported that inflammatory signaling within
the tumor microenvironment affects CSCs (20–23).
Particularly, IL-6 has been reported to induce
epithelial-mesenchymal transition, which has been implicated in the
generation of a stem cell phenotype (20,21). The
main inflammatory cells in the tumor microenvironment are
lymphocytes and macrophages, and the main inflammatory cytokines
include TNF-α, IL-6, IL-8 and IFN-γ. Based on the results of
previous in vitro studies (20–23), the
association between inflammation and CSCs in human breast cancer
tissue was analyzed in the present study. The results of the
current study demonstrated that intratumoral inflammation and
tumor-infiltrating CD4+ T cell counts are associated
with CSCs in breast cancer. Typically, activated Th1 cells secrete
TNF-α, IL-2, TGF-β and IFN-γ, and activated Th2 cells secrete IL-4,
−5, −6, −10 and −13 (24,25). In combination with the results of
previous studies, the results of the current study suggest that
tumor-infiltrating lymphocytes are implicated in the generation of
CSCs through their secretion of inflammatory cytokines.
It has been suggested that CSCs mediate tumor growth
and metastasis (8,10). However, the prognostic significance of
CSCs in breast cancer remains unclear. Previous studies have
reported that BCSCs are associated with the basal-like molecular
subtype of breast cancer and a poor clinical outcome (26,27).
However, Mylona et al (28)
revealed that BCSCs are associated with a lack of lymph node
metastasis and an improved clinical outcome. Furthermore, Abraham
et al (29) reported that
BCSCs were not associated with the clinical outcome. Notably,
consistent with these previous studies, the results of the present
study demonstrated that a BCSC phenotype
(CD44+/CD24−) was significantly associated
with the basal-like molecular subtype of breast cancer, which
confers a poor prognosis, whereas it was significantly inversely
associated with lymph node metastasis. These results suggest that
BCSCs may be able to initiate tumorigenesis (30), but that signaling pathways that
modulate BCSCs, and interactions between BCSCs and the tumor
microenvironment, may affect breast cancer progression. Further
studies are required to clarify the prognostic significance of CSC
phenotype in breast cancer.
It has been recognized that inflammatory mediators
in the tumor microenvironment affect breast cancer development and
progression (3–7,31).
Previous studies have revealed that cytotoxic T lymphocytes and
natural killer cells exhibit antitumor activity against breast
cancer (32–34). However, numerous studies (31,35–37) have
demonstrated mechanisms by which breast tumors avoid antitumor
immune responses; protumorigenic inflammation in breast cancer has
been reported (31). The inflammatory
mediators in breast carcinogenesis include proinflammatory
cytokines and chemokines, including IL-1, IL-6, IL-8, TNF-α, MCP-1,
CCL5 and CXCL1/2 (3,6,7).
Furthermore, previous studies have suggested that CD8+ T
cells exhibit antitumor activity in breast cancer, which is
dependent on the breast cancer subtype (38,39). Liu
et al (38) reported that
CD8+ T cell infiltration was associated with improved
patient survival in basal-like, but not non-basal, triple negative
breast cancer. In the current study, the clinicopathological
significance of inflammatory mediators and inflammatory cells were
investigated, and tumor-infiltrating CD8+ T cells were
revealed to be increased in patients with the basal-like subtype of
breast cancer compared with other subtypes. However, it was not
possible to analyze the prognostic role of tumor-infiltrating
CD8+ T cells in basal-like breast cancer in the current
study. Further studies investigating the mechanism by which
inflammation influences the progression of different breast cancer
subtypes are warranted.
Although preliminary evidence suggests that
inflammation and BCSCs are associated with breast carcinogenesis,
there is limited data available. The acquisition of more clinical
evidence is important for designing effective therapies and
identifying improved therapeutic targets for patients with breast
cancer. The present study analyzed the association between
inflammation and the BCSC phenotype in human breast cancer tissue.
However, the results of the current study were limited due to a
relatively small sample size, and the clinical significance of
these results requires further evaluation.
In conclusion, the present study identified
significant associations between inflammation and the BCSC
phenotype in breast cancer. The results suggest that the
interaction between inflammation and BCSCs may affect
tumorigenesis, in addition to the progression of breast cancer.
Further studies are required to clarify the role of inflammation
and BCSCs in breast cancer.
Acknowledgements
The authors wish to thank Young Chae Chang (Research
Institute of Biomedical Engineering and Department of Medicine,
Catholic University of Daegu School of Medicine, Daegu, Korea) and
Hyun Ji Cho (Research Institute of Biomedical Engineering and
Department of Medicine, Catholic University of Daegu School of
Medicine, Daegu, Korea) for their technical support and
interpretation of data. The present study was supported by a grant
from the Daegu-Gyeongbuk Surgical Society research foundation,
Korea.
References
|
1
|
Balkwill F and Mantovani A: Inflammation
and cancer: Back to Virchow? Lancet. 357:539–545. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arias JI, Aller MA and Arias J: Cancer
cell: Using inflammation to invade the host. Mol Cancer. 6:292007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Soria G, Ofri-Shahak M, Haas I,
Yaal-Hahoshen N, Leider-Trejo L, Leibovich-Rivkin T, Weitzenfeld P,
Meshel T, Shabtai E, Gutman M and Ben-Baruch A: Inflammatory
mediators in breast cancer: Coordinated expression of TNF-α &
IL-1β with CCL2 & CCL5 and effects on epithelial-to-mesenchymal
transition. BMC Cancer. 11:1302011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lewis CE and Hughes R: Inflammation and
breast cancer. Microenvironmental factors regulating macrophage
function in breast tumours: Hypoxia and angiopoietin-2. Breast
Cancer Res. 9:2092007. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin EY and Pollard JW: Tumor-associated
macrophages press the angiogenic switch in breast cancer. Cancer
Res. 67:5064–5066. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Soria G and Ben-Baruch A: The inflammatory
chemokines CCL2 and CCL5 in breast cancer. Cancer Lett.
267:271–285. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Goldberg JE and Schwertfeger KL:
Proinflammatory cytokines in breast cancer: Mechanisms of action
and potential targets for therapeutics. Curr Drug Targets.
11:1133–1146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Korkaya H, Kim GI, Davis A, Malik F, Henry
NL, Ithimakin S, Quraishi AA, Tawakkol N, D'Angelo R, Paulson AK,
et al: Activation of an IL6 inflammatory loop mediates trastuzumab
resistance in HER2+ breast cancer by expanding the
cancer stem cell population. Mol Cell. 47:570–584. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Iqbal J, Chong PY and Tan PH: Breast
cancer stem cells: An update. J Clin Pathol. 66:485–490. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Clarke MF, Dick JE, Dirks PB, Eaves CJ,
Jamieson CH, Jones DL, Visvader J, Weissman IL and Wahl GM: Cancer
stem cells-perspectives on current status and future directions:
AACR Workshop on cancer stem cells. Cancer Res. 66:9339–9344. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Boyle ST and Kochetkova M: Breast cancer
stem cells and the immune system: Promotion, evasion and therapy. J
Mammary Gland Biol Neoplasia. 19:203–211. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gammaitoni L, Leuci V, Mesiano G, Giraudo
L, Todorovic M, Carnevale-Schianca F, Agiletta M and Sangiolo D:
Immunotherapy of cancer stem cells in solid tumors: Initial
findings and future prospective. Expert Opin Biol Ther.
14:1259–1270. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
AJCC Cancer Staging Manual, . 7th edition.
Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL and Trotti FL:
Springer-Verlag; New York: pp. 347–377. 2010
|
|
14
|
Elston CW and Ellis IO: Pathological
prognostic factors in breast cancer I. The value of histological
grade in breast cancer: Experience from a large study with
long-term follow-up. Histopathology. 19:403–410. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jeong YJ, Jeong HY, Bong JG, Park SH and
Oh HK: Low methylation levels of the SFRP1 gene are associated with
the basal-like subtype of breast cancer. Oncol Rep. 29:1946–1954.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brabletz T: EMT and MET in metastasis:
Where are the cancer stem cells? Cancer Cell. 22:699–701. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shigdar S, Li Y, Bhattacharya S, O'Connor
M, Pu C, Lin J, Wang T, Xiang D, Kong L, Wei MQ, et al:
Inflammation and cancer stem cells. Cancer Lett. 345:271–278. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vermeulen L, De Sousa Melo E, van der
Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M,
Merz C, Rodermond H, et al: Wnt activity defines colon cancer stem
cells and is regulated by the microenvironment. Nat Cell Biol.
12:468–476. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Blaylock RL: Cancer microenvironment,
inflammation and cancer stem cells: A hypothesis for a paradigm
change and new targets in cancer control. Surg Neurol Int.
6:922015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iliopoulos D, Hirsch HA, Wang G and Struhl
K: Inducible formation of breast cancer stem cells and their
dynamic equilibrium with non-stem cancer cells via IL6 secretion.
Proc Natl Acad Sci USA. 108:1397–1402. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sullivan NJ, Sasser AK, Axel AE, Vesuna F,
Raman V, Ramirez N, Oberyszyn TM and Hall BM: Interleukin-6 induces
an epithelial-mesenchymal transition phenotype in human breast
cancer cells. Oncogene. 28:2940–2947. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sansone P, Storci G, Tavolari S, Guarnieri
T, Giovannini C, Taffurelli M, Ceccarelli C, Santini D, Paterini P,
Marcu KB, et al: IL-6 triggers malignant features in mammospheres
from human ductal breast carcinoma and normal mammary gland. J Clin
Invest. 117:3988–4002. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Katanov C, Lerrer S, Liubomirski Y,
Leider-Trejo L, Meshel T, Bar J, Feniger-Barish R, Kamer I,
Soria-Artzi G, Kahani H, et al: Regulation of the inflammatory
profile of stromal cells in human breast cancer: Prominent roles
for TNF-α and the NF-κB pathway. Stem Cell Res Ther. 6:872015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
DeNardo DG and Coussens LM: Inflammation
and breast cancer. Balancing immune response: Crosstalk between
adaptive and innate immune cells during breast cancer progression.
Breast Cancer Res. 9:2122007. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Goto S, Sato M, Kaneko R, Itoh M, Sato S
and Takeuchi S: Analysis of Th1 and Th2 cytokine production by
peripheral blood mononuclear cells as a parameter of immunological
dysfunction in advanced cancer patients. Cancer Immunol Immunother.
48:435–442. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Idowu MO, Kmieciak M, Dumur C, Burton RS,
Grimes MM, Powers CN and Manjili MH: CD44(+)/CD24(−/low) cancer
stem/progenitor cells are more abundant in triple-negative invasive
breast carcinoma phenotype and are associated with poor outcome.
Hum Pathol. 43:364–373. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim HJ, Kim MJ, Ahn SH, Son BH, Kim SB,
Ahn JH, Noh WC and Gong G: Different prognostic significance of
CD24 and CD44 expression in breast cancer according to hormone
receptor status. Breast. 20:78–85. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mylona E, Giannopoulou I, Fasomytakis E,
Nomikos A, Magkou C, Bakarakos P and Nakopoulou L: The
clinicopathologic and prognostic significance of
CD44(+)/CD24(−/low) and CD44−/CD24+ tumor
cells in invasive breast carcinomas. Hum Pathol. 39:1096–1102.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Abraham BK, Fritz P, McClellan M,
Hauptvogel P, Athelogou M and Brauch H: Prevalence of
CD44+/CD24−/low cells in breast cancer may
not be associated with clinical outcome but may favor distant
metastasis. Clin Cancer Res. 11:1154–1159. 2005.PubMed/NCBI
|
|
30
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad USA. 100:3983–3988.
2003. View Article : Google Scholar
|
|
31
|
Jiang X and Shapiro DJ: The immune system
and inflammation in breast cancer. Mol Cell Endocrinol.
382:673–682. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tkach M, Coria L, Rosemblit C, Rivas MA,
Proietti CJ, Díaz Flaqué MC, Beguelin W, Frahm I, Charreau EH,
Cassataro J, et al: Targeting Stat3 induces senescence in tumor
cells and elicits prophylactic and therapeutic immune responses
against breast cancer growth mediated by NK cells and
CD4+ T cells. J Immunol. 189:1162–1172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang B, Zaidi N, He LZ, Zhang L, Kuroiwa
JM, Keler T and Steinman RM: Targeting of the non-mutated tumor
antigen HER2/neu to mature dendritic cells induces an integrated
immune response that protects against breast cancer in mice. Breast
Cancer Res. 14:R392012. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mamessier E, Sylvain A, Thibult ML,
Houvenaeghel G, Jacquemier J, Castellano R, Gonçalves A, André P,
Romagné F, Thibault G, et al: Human breast cancer cells enhance
self tolerance by promoting evasion from NK cell antitumor
immunity. J Clin Invest. 121:3609–3622. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tsukerman P, Stern-Ginossar N, Gur C,
Glasner A, Nachmani D, Bauman Y, Yamin R, Vitenshtein A, Stanietsky
N, Bar-Mag T, et al: MiR-10b downregulates the stress-induced cell
surface molecule MICB, a critical ligand for cancer cell
recognition by natural killer cells. Cancer Res. 72:5463–5472.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jiang X, Ellison SJ, Alarid ET and Shapiro
DJ: Interplay between the levels of estrogen and estrogen receptor
controls the level of the granzyme inhibitor, proteinase inhibitor
9 and susceptibility to immune surveillance by natural killer
cells. Oncogene. 26:4106–4114. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bargou RC, Wagener C, Bommert K, Mapara
MY, Daniel PT, Arnold W, Dietel M, Guski H, Feller A, Royer HD and
Dorken B: Overexpression of the death-promoting gene bax-alpha
which is downregulated in breast cancer restores sensitivity to
different apoptotic stimuli and reduces tumor growth in SCID mice.
J Clin Invest. 97:2651–2659. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu S, Lachapelle J, Leung S, Gao D,
Foulkes WD and Nielsen TO: CD8+ lymphocyte infiltration
is an independent favorable prognostic indicator in basal-like
breast cancer. Breast Cancer Res. 14:R482012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mahmoud SM, Paish EC, Powe DG, Macmillan
RD, Grainge MJ, Lee AH, Ellis IO and Green AR: Tumor-infiltrating
CD8+ lymphocytes predict clinical outcome in breast
cancer. J Clin Oncol. 29:1949–1955. 2011. View Article : Google Scholar : PubMed/NCBI
|