Introduction
Malignant melanoma is one of the most aggressive
forms of cancer, exhibiting resistance to various forms of
chemotherapy. The global incidence and mortality rates of malignant
melanoma are increasing (1–3). Novel targeted therapies designed to kill
melanoma cells harboring mutations in B-Raf proto-oncogene,
serine/threonine kinase (BRAF) have been developed using
vemurafenib, which is a specific BRAF inhibitor (4,5). Missense
mutations to the BRAF gene, most commonly a valine-to-glutamic acid
substitution at codon 600, has been observed in ~80% of melanocytic
nevi and ~50% of melanomas (6–9). In
addition to this BRAF mutation, another therapeutic target has been
investigated, as melanomas, including acral lentiginous melanoma,
the most common type in ethnicities that produce high levels of
melanin, have a low frequency of BRAF gene mutation (10,11).
However, details of the mechanism responsible for the drug
insensitivity of melanomas lacking BRAF mutations have been largely
unclear. On the basis of the aforementioned background, a search
for novel therapeutic strategies is justified.
β-lapachone (β-lap), a lipophilic cytotoxic
o-naphtoquinone derived from the bark of the South American
Lapacho tree (Tabebuia avellanedae), has recently attracted
attention as an antitumor drug (12,13). The
mechanism of its antitumor effect is considered to involve the
formation of reactive oxygen species (ROS) (13–15). ROS
and other types of radicals are involved in a variety of biological
phenomena, including tumorigenesis, degenerative disease and aging
(16,17), and are also important mediators of
tumor cell death (18). NAD(P)H
quinone oxidoreductase 1 (NQO1) catalyzes the reduction of β-lap to
an unstable hydroquinone, which then undergoes rapid oxidization
and is reconverted to a stable quinone (14). This repeated oxidation-reduction cycle
induces ROS, partially contributing to the tumor killing activity
of β-lap (14,15,18).
Under physiological conditions, the intracellular
level of ROS is tightly regulated by NF-E2 related factor 2
(NFE2L2, also known as NRF2) and its inhibitor protein, Kelch-like
ECH-associated protein 1 (KEAP1), which mediates NRF2 degradation.
NRF2 is a transcription factor that forms a heterodimer with one of
the small Maf-family proteins and binds to an
antioxidant-responsive element to activate transcription of target
genes, including NQO1 (19,20).
A previous study revealed that several melanoma cell
lines have detectable endogenous expression of NQO1 (21). Certain patients with melanoma have a
mutation at the KEAP1 locus, which results in NRF2 stabilization
(22). On the basis of these
findings, we hypothesized that certain melanoma cells may be
constitutionally sensitive to NQO1-dependent antitumor drugs,
including β-lap, and that this sensitivity may be further increased
through activation of the KEAP1-NRF2 axis.
The present study investigated whether forced
induction of NQO1, through NRF2 activation, sensitizes melanoma
cells to β-lap. First, whether NQO1 mediated β-lap toxicity in
melanoma cell lines was assessed. Next, to achieve overexpression
of NQO1, phytochemical carnosic acid (CA), which is a potent
activator of the KEAP1/NRF2 system, was used (23). The combined administration of CA
augmented the antitumor effect of β-lap in several melanoma cell
lines. The findings of the present study indicate the potential
availability of β-lap and CA in combination as a novel
chemotherapeutic approach for malignant melanoma.
Materials and methods
Cell cultures
Various human melanoma cell lines were obtained from
the following sources: CRL-1585 (also known as C32 cells), G-361,
HMV-II and SK-MEL-28 from the Cell Resource Center for Biomedical
Research, Tohoku University (Sendai, Japan); MeWo, SK-MEL-2 and
SK-MEL-31 from the American Type Culture Collection (ATCC;
Manassas, VA, USA); MM-AN were provided by Dr M. C. Mihm
(Department of Dermatology, Harvard Medical School, Boston, MA,
USA); GAK and HMY-1 from the Japanese Collection of Research
Bioresources (Osaka, Japan). The murine melanoma B16BL6 cell line
was also obtained from the RIKEN BioResource Center (Tsukuba,
Japan). The human non-small cell lung cancer H460 cell line was
also obtained from ATCC. The identities of cell lines used in this
study were confirmed by a short tandem repeat analysis (data not
shown). The cells were maintained at 37°C under 5% CO2
in RPMI-1640 supplemented with 10% fetal bovine serum (FBS) and 1%
penicillin-streptomycin (all from Thermo Fisher Scientific, Inc.,
Waltham, MA, USA).
Small interfering RNA (siRNA)
transfection
Silencer select siRNAs against NQO1 (cat. no.
4390824; IDs, s4089, s4090, and s4091), NRF2 (cat. no. 4392420; ID,
s9491), and a negative control siRNA (cat. no. 4390844),
Lipofectamine RNAiMAX transfection reagent and Opti-MEM were all
obtained from Thermo Fisher Scientific, Inc. Silencer Select siRNAs
were pre-designed and validated by the manufacturer. Cells at 50%
confluence were treated for 72 h in prior to the subsequent
experiments, with 10 nM siRNA, 10 µl Lipofectamine RNAiMAX
transfection reagent, 1 ml Opti-MEM and 9 ml RPMI-1640 supplemented
with 10% FBS and 1% penicillin-streptomycin in a 10-cm dish plate
in accordance with the manufacturer's instructions.
Reagents and antibodies
β-lap, carnosic acid (CA), and a NQO1 inhibitor,
ES936, were obtained from Sigma-Aldrich; Merck KGaA (Darmstadt,
Germany). Stock solutions were prepared by dissolving the chemicals
in dimethyl sulfoxide at 20 mM (β-lap), 100 mM (CA), and 10 mM
(ES936).
The antibody directed against NRF2 (cat. no.
ab-62352) was obtained from Abcam (Cambridge, MA, USA). The
antibody against NQO1 (cat. no. 3187) was obtained from Cell
Signaling Technology, Inc. (Danvers, MA, USA). An antibody against
β-actin (cat. no. A2228) was obtained from Sigma-Aldrich; Merck
KgaA. Anti-Rabbit IgG, HRP-Linked F(ab')2 Fragment
Donkey (cat. no. NA9340) and Anti-Mouse IgG, HRP-Linked Whole Ab
Sheep (cat. no. NA931) were obtained from GE Healthcare (Chicago,
IL USA). The fluorescent dyes against rabbit IgG (Alexa Fluor-488)
was obtained from Thermo Fisher Scientific, Inc. The fluorescent
dyes against mouse IgG (Alexa Fluor-594) was obtained from Thermo
Fisher Scientific, Inc. DAPI was obtained from Dojindo Molecular
Technologies, Inc. (Kumamoto, Japan).
Cell viability assay
A Cell Counting kit-8 (Dojindo Molecular
Technologies, Inc.), which utilizes water-soluble tetrazolium
salts, was used to evaluate the proliferation of melanoma cells
following drug treatment. All melanoma cell lines were seeded into
96-well plates (5,000 cells/well) and cultured for 24 h prior to
treatment with β-lap and/or CA. Following the treatment, the medium
in each well was replaced with 100 µl of drug-free fresh medium and
10 µl of Cell Counting kit-8 solution, incubated for an additional
1–2 h, and the absorbance of each well at 450 nm was measured using
a Multiskan Spectrum spectrophotometer (Thermo Fisher Scientific,
Inc.).
Western blotting
All melanoma cell lines at 80–90% confluence, which
were maintained at 37°C under 5% CO2 in RPMI-1640
supplemented with 10% FBS and 1% penicillin-streptomycin, were
washed twice with ice-chilled PBS, treated with 10% trichloroacetic
acid for 30 min on ice and then scraped off into a tube. The cell
pellet was washed once with deionized water and lysed in 9 M urea,
2% Triton X-100, and 1% dithiothreitol (DTT). Protein concentration
was measured using a BCA protein assay kit (EMD Millipore,
Billerica, MA, USA) prior to the addition of DTT. Protein samples
were separated using SDS-PAGE (10% gel) and then transferred onto
polyvinylidene difluoride transfer membranes (Pall Corporation,
Portsmouth, UK). All proteins were loaded 30 µg/lane.
For all antibodies, the membranes were blocked with
5% non-fat dried milk in 0.1% Tween-20/PBS at room temperature for
1 h, then probed with an appropriate primary antibodies overnight
at 4°C, which were diluted to 1:1,000, and with HRP-conjugated
secondary antibodies for 1 h at room temperature, which were
diluted to 1:5,000. Each antibody was diluted in 5% non-fat dried
milk in 0.1% PBS-Tween-20. Signals were detected with ECL prime
detection reagents (GE Healthcare) and ChemiDoc XRS (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Densitometric analysis of
each protein signal was performed using ImageJ software (version
1.50; National Institutes of Health, Bethesda, MD, USA) (24).
Auxin-inducible degron (AID)
system
The B16BL6/pAO1 cell line, conditionally expressing
NQO1 under the control of the AID system, was previously
established (25). B16BL6/pAO1 cells
were treated with β-lap and/or 0.5 mM auxin (BioROIS Co., Ltd.,
Mishima, Japan) for 24 h in 96-well plates. Cell viability was
measured using a CCK-8 (Dojindo Molecular Technologies, Inc.)
accordng to the manufacturer's instructions. To detect NQO1 using
immunoblotting, B16BL6/pAO1 cells were treated with or without 0.5
mM auxin for 24 h in 10-cm dish plates. Following this treatment,
western blotting was performed as aforementioned.
Immunofluorescence staining
Cells were washed with PBS and fixed with 4.0%
formaldehyde and 0.5% Triton X-100 (Sigma-Aldrich; Merck KGaA) for
20 min at room temperature. The slides were then blocked using 5%
FBS (Thermo Fisher Scientific, Inc.) and 0.5% Triton X-100 for 30
min at room temperature. Following three washes with PBS, the
slides were incubated with a primary antibody (NRF2 or NQO1) in
blocking buffer (PBS with 5% FBS and 0.5% Triton X-100) at 4°C,
which were diluted to 1:500. The slides were incubated with
secondary antibodies: Alexa Fluor-488 (cat. no. A11008) and Alexa
Fluor-594 (cat. no. A11005) (1:250; both from Thermo Fisher
Scientific, Inc.) for 3 h at 37°C. Cells were counterstained with
DAPI (Dojindo Molecular Technologies, Inc.) for 30 min at room
temperature and images were captured using a KEYENCE BZ9000
fluorescence microscope (Keyence Corporation, Osaka, Japan). Images
were captured at magnification, ×10.
Statistical analysis
Pearson's correlation coefficient was used to assess
the correlation between cell viability and relative NQO1 protein
expression. In the cell viability assay, a t-test with the
Bonferroni correction was conducted on raw data for the respective
doses. P<0.05 was considered to indicate a statistically
significant difference. Fitting to a sigmoid function was performed
for estimating the half-maximal inhibitory concentration
(IC50). One-way analysis of variance followed by Tukey's
honest significant difference post-hoc test was used to assess the
effect of CA on NQO1 expression. All analyses were conducted using
Microsoft Excel 2013 (Microsoft Corporation, Redmond, WA, USA) and
RStudio Desktop version 1.0.136 (R Studio, Boston, MA, USA).
Results
Effect of β-lap on melanoma cell
lines
A previous study demonstrated that melanoma cell
lines exhibit detectable endogenous expression of NQO1, with the
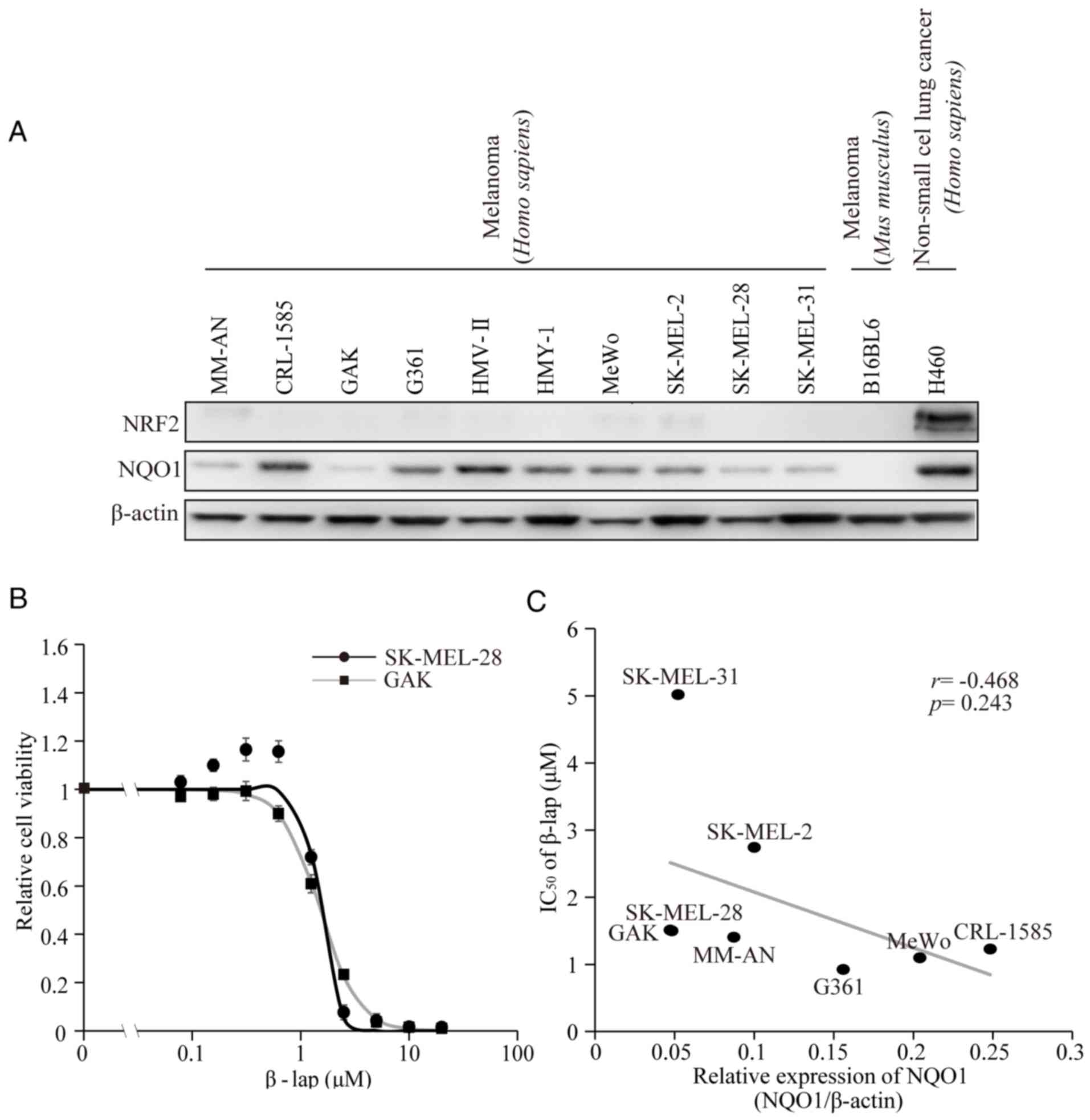
level of expression varying among the cell lines (21). In the present study, the protein
expression levels of NQO1 and NRF2 were verified (Fig. 1A). As it has been shown that NQO1 is
required for the bio-activation of β-lap (14), it was hypothesized that melanoma cells
may be sensitive to the anti-proliferative effect of β-lap. To
confirm the effect of β-lap, cell viability was preliminarily
measured following treatment of the melanoma SK-MEL-28 and GAK cell
lines with various concentrations of β-lap. The two cell lines
exhibited dose-dependent decreases of cell viability (Fig. 1B). The calculated IC50
values for β-lap were 1.49 and 1.51 µM for SK-MEL-28 and GAK cells,
respectively (Table I). The
viabilities of the remaining six cell lines were also tested and
the IC50 values are presented in Table I. These corresponded to the values
that had been reported previously for SK-MEL-28 and another cell
line, G361 (26). The association
between the basal expression levels of NQO1 and the IC50
values of β-lap in eight melanoma cell lines were then compared.
Differences in the basal NQO1 expression level were detected
(Fig. 1C and Table I). However, no significant correlation
between the basal NQO1 expression levels and IC50 values
of β-lap was observed (R=−0.468, P=0.243).
 | Table I.Effect of CA or ES936 on sensitivity
of β-lap in melanoma cell lines. |
Table I.
Effect of CA or ES936 on sensitivity
of β-lap in melanoma cell lines.
|
| IC50 of
β-lap, µM |
|
|
|---|
|
|
|
|
|
|---|
|
|
| CA |
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Cell line | Control | 0.5 µM | 40 µM | ES936 | NQO1/β-actin | Fold increase of
NQO1 by 40 µM CA |
|---|
| MM-AN | 1.41 | 1.16 | 0.42 | 4.77 | 0.087 | 1.25 |
| CRL-1585 | 1.23 | 0.95 | 0.28 | 5.70 | 0.248 | 1.26 |
| GAK | 1.51 | 1.38 | 0.71 | 2.70 | 0.047 | 1.53 |
| G361 | 0.93 | 0.77 | 0.14 | 3.80 | 0.156 | 1.28 |
| MeWo | 1.20 | 0.80 | 0.30 | 2.70 | 0.204 | 1.85 |
| SK-MEL-2 | 2.74 | 1.91 | 0.73 | 4.63 | 0.100 | 1.32 |
| SK-MEL-28 | 1.49 | 1.38 | 0.62 | 9.15 | 0.048 | 1.63 |
| SK-MEL-31 | 5.02 | 2.54 | 2.05 | 11.72 | 0.052 | 1.32 |
NQO1-dependent cytotoxicity of β-lap
in melanoma cell lines
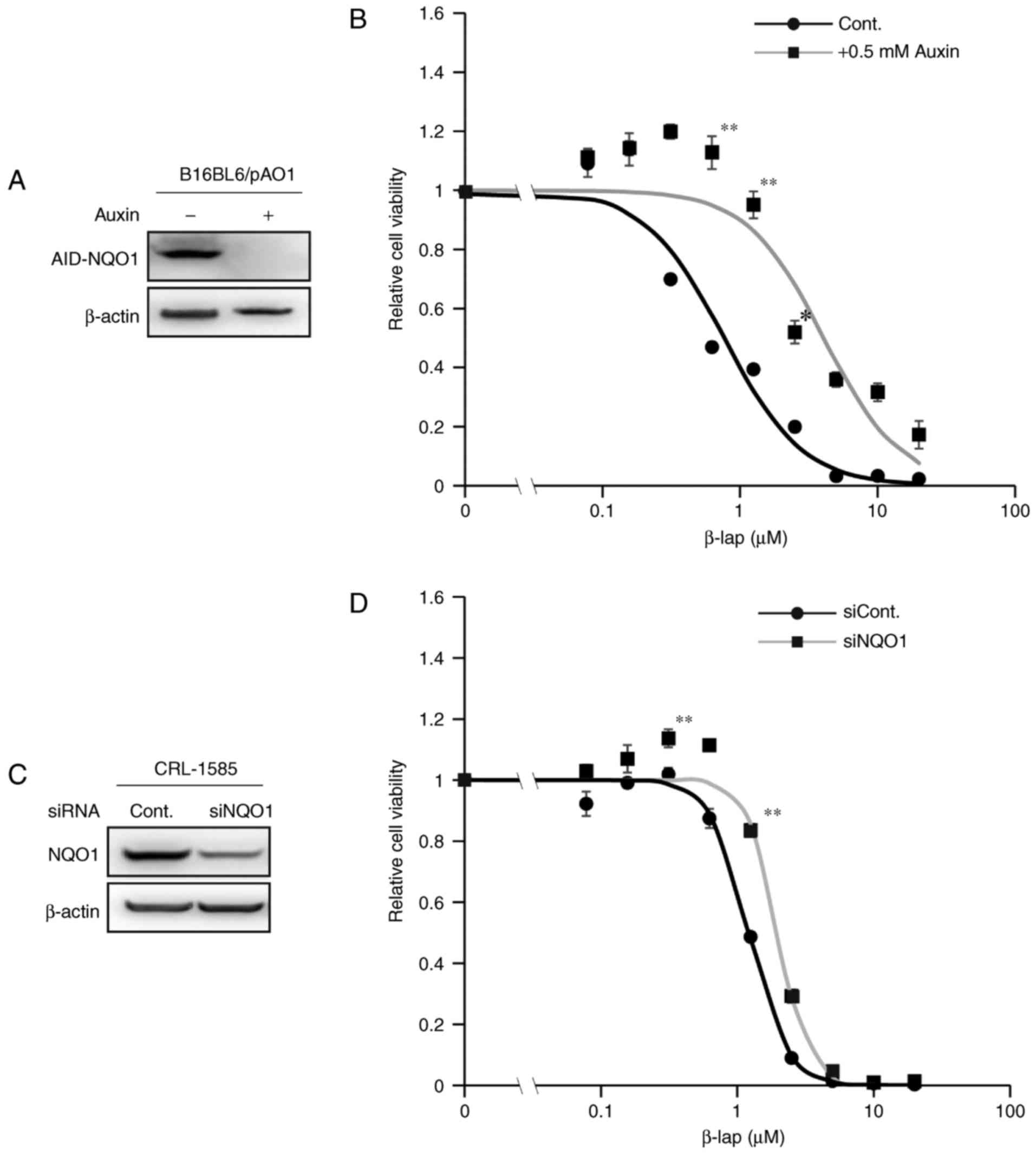
To assess whether NQO1 is involved in β-lap-mediated
toxicity in melanoma cell lines, a cell line that conditionally
expresses NQO1 under the control of the AID system was used
(25,27). In this system, addition of the plant
hormone auxin induces the rapid ubiquitination of AID, followed by
the proteosome-mediated degradation of ectopically expressed
AID-fused NQO1 within 1 h (Fig. 2).
As presented in Fig. 2B,
β-lap-mediated toxicity in melanoma cells was decreased following
auxin treatment in comparison with untreated controls, as assessed
by a cell viability assay. For the experiment using siRNAs,
CRL-1585 cells were selected as they have relatively higher
endogenous expression of NQO1 compared with the other cell lines
(Fig. 1A and C). The β-lap-mediated
toxicity was decreased following treatment with siRNA against NQO1
(Fig. 2D). However, the effect was
not as evident as that in the AID system (Fig. 2A and B), potentially because
endogenous NQO1 expression was not completely abolished by
transfection with the NQO1 siRNA (Fig.
2C). These results indicated that NQO1 expression was involved
in the cytotoxicity of β-lap in melanoma cell lines.
CA stabilizes NRF2 and induces further
expression of NQO1
CA is a potent activator of the KEAP1-NRF2 axis
(23). In the absence of CA, NRF2 is
degraded through the formation of a complex with its inhibitor
protein, KEAP1, a member of the E3 ligase family (20). CA induces conformational changes by
targeting the cysteine residues on KEAP1 proteins via thiol
S-alkylation and the released NRF2 is stabilized by escaping from
its degradation complex (23,28). NRF2 is then able to translocate into
the nucleus and operate as a transcription factor, forming a
heterodimer with one of the small Maf-family proteins, and binding
to an antioxidant-responsive element to activate the transcription
of target genes, for instance NQO1 (20). Therefore, using the melanoma SK-MEL-28
and GAK cell lines, whether CA treatment would stabilize NRF2 was
assessed, leading to further induction of NQO1 expression. The two
cell lines had low basal NQO1 and NRF2 expression (Fig. 1A) compared with the non-small cell
lung cancer H460 cell line, which has been shown to harbor somatic
mutations in the KEAP1 gene, resulting in the high expression of
NRF2 (29). Concentrations of CA that
did not affect cell viability were used (Fig. 3A and B). Notably, CA was less toxic
compared with other KEAP1-NRF2 activators, including sulforaphane
(Fig. 3A and B). The expression of
NRF2 was associated with that of NQO1 in the two melanoma cell
lines (Fig. 3C and D). Furthermore,
to confirm the activation of the KEAP1-NRF2 axis by CA in melanoma
cell lines, western blot analysis was performed to assess the
amount of NRF2 and NQO1 in siRNA-transfected melanoma cell lines.
As expected, it was revealed that the significant increase in NQO1
induced by CA was abolished in siNRF2-treated melanoma cell lines
(Fig. 3C and D). Furthermore, as
assessed by immunofluorescence staining, CA treatment resulted in
further NRF2 stabilization and induction of NQO1 expression in the
entire population of the two cell lines (Fig. 3E and F). These data indicate that CA
may be used as an inducer of NQO1 in melanoma cell lines.
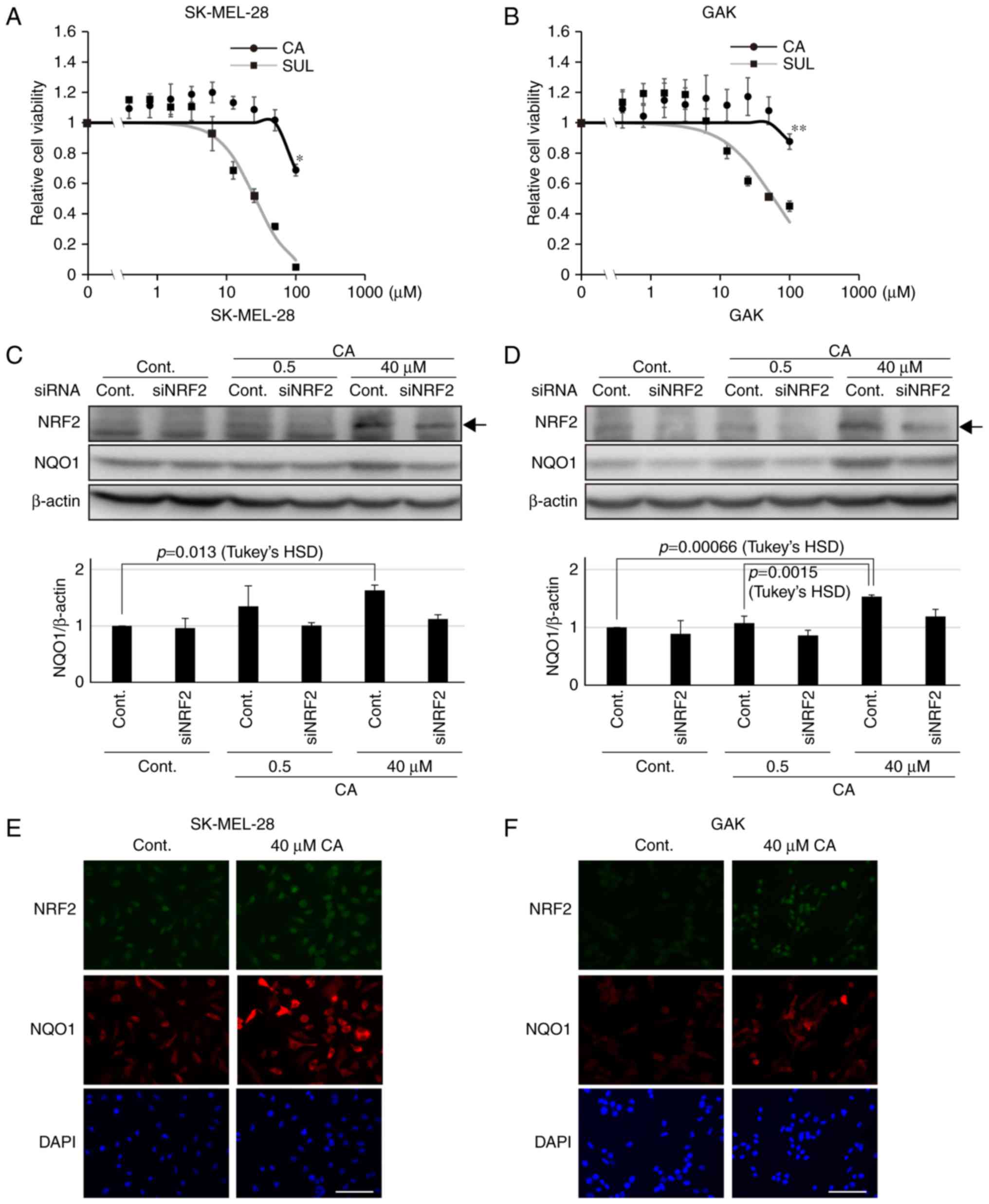 | Figure 3.Carnosic acid stabilizes NRF2 and
induces further expression of NQO1. Viability of (A) SK-MEL-28 and
(B) GAK cells treated for 24 h with CA or SUL. One-way ANOVA
followed by Tukey's honest significant difference test was used for
statistical analysis (*P<0.05; **P<0.01). The expression of
NRF2 and NQO1 determined by western blotting in siNRF2-treated (C)
SK-MEL-28 or (D) GAK cells in the presence of CA at 0.5 or 40 µM
for 24 h. Densitometrically quantified expression levels of
NQO1/β-actin are shown below. Arrows indicate the band of NRF2.
Bars in the graphs indicate the mean ± standard error of the mean
of three independent experiments. In SK-MEL-28 and GAK cells, the
effect of CA on NQO1 expression was significant in the control
series (P=0.016 and P=0.000538 by one-way ANOVA, respectively) and
not so in siNRF2 series (P=0.571 and P=0.34, respectively).
Immunofluorescent staining of NRF2 and NQO1 in (E) SK-MEL-28 or (F)
GAK cells treated with 40 µM of CA for 24 h. Scale bar, 100 µm.
siRNA, small interfering RNA; NRF2, NF-E2 related factor 2; NQO1,
NAD(P)H quinone oxidoreductase 1; ANOVA, analysis of variance; CA,
carnosic acid; SUL, suforaphane; cont, control. |
Combination treatment with CA
increases the sensitivity of melanoma cells to β-lap
Next, whether the induction of the expression of
NQO1 was able to increase the sensitivity of melanoma cell lines to
β-lap-mediated toxicity was investigated. As presented in Table I, sensitivity to β-lap was increased
by combination treatment with CA. This induction was CA
concentration-dependent (Table I).
Treatment with the specific NQO1 inhibitor ES936 decreased the
sensitivity of all the cell lines to β-lap-mediated toxicity
(Table I). The concentrations of
ES936 used did not affect cell viability (data not shown). These
results indicate that combined treatment with CA increases the
sensitivity of melanoma cells to β-lap through induction of NQO1
expression.
Discussion
Malignant melanoma is one of the most aggressive
types of skin cancer, and its incidence is increasing in Caucasian
and non-Caucasian populations (2,30,31). Although a number of therapeutic
approaches for melanoma have been developed, including
chemotherapy, immunotherapy, surgery and several forms of
molecular-targeted therapy, the response rate of patients has
remained insufficient, and side effects continue to be an issue
(2,30). Therefore, other approaches for the
improvement of treatment outcome requires investigation.
The present study focused on β-lap, a natural
quinone derived from the bark of the Lapacho tree (12,13).
Previous studies have revealed that β-lap acts as a potent
anti-proliferative agent, inhibiting topoisomerase I/II (26,32,33),
specificity protein 1 (26) and the
cell cycle (34). It has also been
shown that repetitive oxidation-reduction of β-lap by NQO1
generates ROS in tumor cells (14),
thus contributing to cytotoxicity in malignancies, including
pancreatic cancer (35,36). Among the various aforementioned
antitumor mechanisms of β-lap, the present study focused on the
role of NQO1 in β-lap-mediated toxicity in melanoma cell lines, as
a previous study had shown that normal melanocytes express higher
levels of NQO1 compared with other tissues (21). As expected, loss or inhibition of NQO1
decreased the degree of β-lap-mediated toxicity. NQO1 is involved
in the regulation of melanin synthesis via suppression of
tyrosinase degradation in melanocytes under physiological
conditions (37). We hypothesized
that the higher basal expression of NQO1 in melanoma cell lines was
due to such a mechanism that is specific to pigmented cells.
Collectively, the results of the present study indicate that
melanomas originating from melanocytes have constitutively high
sensitivity to NQO1-dependent antitumor drugs, including β-lap.
CA is a natural, catechol-type polyphenolic
diterpene derived from rosemary (Rosmarinus officinalis),
comprising about 5% of the dry weight of rosemary leaves (23,38). CA
has various biological effects, mediated via phosphatidylinositol
3-kinase (39), peroxisome
proliferator-activated receptor γ (40), cyclin A/B1 (41) and free radical-scavenging activity
(42). CA is also known to activate
the KEAP1/NRF2 system (23). It
should be emphasized that NQO1 is known to be a typical target gene
of NRF2. The results of the present study demonstrated that CA
treatment led to further induction of NQO1 expression in melanoma
cell lines, at least under the experimental conditions utilized in
the present study. These findings indicate that CA may have a
clinical application as a sensitizer for NQO1-dependent antitumor
drugs; indeed, combination treatment with CA increased the β-lap
sensitivity of all the melanoma cell lines assessed.
Throughout the present study, other than for the
NQO1 knockdown experiment, two cell lines, SK-MEL-28 and GAK, were
used because they exhibited relatively higher induction of NQO1 by
40 µM CA in comparison with another melanoma cell lines.
Furthermore, the induction of NQO1 by CA was more representative
when the levels of NQO1 expression were assessed by western
blotting, as these cell lines have basally express low levels of
NQO1. Notably, NQO1 induction in MeWo cells was the highest among
all cell lines tested. However, this cell line was not selected
owing to its relatively high basal level of NQO1 expression.
Several studies have shown that NQO1 produces β-lap
radicals, leading in turn to generation of the superoxide anion
that stabilizes β-lap (14,15,18). The
present study did not assess direct evidence for the involvement of
radicals produced via NQO1 in the killing of melanoma cell lines.
However, it was demonstrated that the tumor-killing ability of
β-lap was NQO1-dependent, as a decrease in the expression of NQO1
or inhibition by its specific inhibitor ES936, increased the
IC50 value of β-lap. Furthermore, CA treatment induced
the expression of NQO1 in melanoma cell lines. Taken together with
previous studies, the data in the present study indicated that
enhancement of the tumor-killing ability of β-lap by CA may be due
to radicals produced via NQO1.
Thus far, CA has been shown to inhibit cell adhesion
and migration, possibly by reducing the activity of secreted
proteases, including the urokinase plasminogen activator and matrix
metalloproteinases, in several tumor cell lines (43,44). In
addition to the inhibitory effect of CA on cell proliferation, its
effect on cell migration and invasion may represent an attractive
therapeutic option for highly metastatic malignant melanomas.
Combined treatment with β-lap and poly(ADP-ribose) polymerase
(PARP) inhibitors has been shown to exert a synergistic therapeutic
effect in several tumor types by causing non-repairable DNA damage
in the presence of NQO1 activity (45). It is possible that CA treatment may be
able to further augment the combined effect of β-lap and PARP.
An allelic variant of NQO1 with essentially no
enzymatic activity is reported to exist at a high frequency,
particularly in Asian populations (46). A polymorphism of NQO1 has reportedly
been associated with response to chemotherapy (47,48).
Therefore the existing data indicated that confirmation of genetic
background prior to β-lap treatment would be warranted for patients
with malignant melanoma.
As β-lap and CA appear to have few side effects, the
results of the present study support the possibility that their use
in combination to increase the expression of NQO1 may provide a
novel avenue of treatment for patients with malignant melanoma.
References
|
1
|
Scolyer RA, Judge MJ, Evans A, Frishberg
DP, Prieto VG, Thompson JF, Trotter MJ, Walsh MY, Walsh NM and
Ellis DW: International Collaboration on Cancer Reporting: Data set
for pathology reporting of cutaneous invasive melanoma:
Recommendations from the international collaboration on cancer
reporting (ICCR). Am J Surg Pathol. 37:1797–1814. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tokuzumi A, Fukushima S, Miyashita A,
Nakahara S, Kubo Y, Yamashita J, Harada M, Nakamura K, Kajihara I,
Jinnin M and Ihn H: Cell division cycle-associated protein 1 as a
new melanoma-associated antigen. J Dermatol. 43:1399–1405. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bollag G, Hirth P, Tsai J, Zhang J,
Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, et al:
Clinical efficacy of a RAF inhibitor needs broad target blockade in
BRAF-mutant melanoma. Nature. 467:596–599. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tsai J, Lee JT, Wang W, Zhang J, Cho H,
Mamo S, Bremer R, Gillette S, Kong J, Haass NK, et al: Discovery of
a selective inhibitor of oncogenic B-Raf kinase with potent
antimelanoma activity. Proc Natl Acad Sci USA. 105:3041–3046. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W,
et al: Mutations of the BRAF gene in human cancer. Nature.
417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Maldonado JL, Fridlyand J, Patel H, Jain
AN, Busam K, Kageshita T, Ono T, Albertson DG, Pinkel D and Bastian
BC: Determinants of BRAF mutations in primary melanomas. J Natl
Cancer Inst. 95:1878–1890. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pollock PM, Harper UL, Hansen KS, Yudt LM,
Stark M, Robbins CM, Moses TY, Hostetter G, Wagner U, Kakareka J,
et al: High frequency of BRAF mutations in nevi. Nat Genet.
33:19–20. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Uribe P, Wistuba II and González S: BRAF
mutation: A frequent event in benign, atypical and malignant
melanocytic lesions of the skin. Am J Dermatopathol. 25:365–370.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bradford PT, Goldstein AM, McMaster ML and
Tucker MA: Acral lentiginous melanoma: Incidence and survival
patterns in the United States, 1986–2005. Arch Dermatol.
145:427–434. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kogushi-Nishi H, Kawasaki J, Kageshita T,
Ishihara T and Ihn H: The prevalence of melanocytic nevi on the
soles in the Japanese population. J Am Acad Dermatol. 60:767–771.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Planchon SM, Wuerzberger S, Frydman B,
Witiak DT, Hutson P, Church DR, Wilding G and Boothman DA:
Beta-lapachone-mediated apoptosis in human promyelocytic leukemia
(HL-60) and human prostate cancer cells: A p53-independent
response. Cancer Res. 55:3706–3711. 1995.PubMed/NCBI
|
|
13
|
Docampo R, Cruz FS, Boveris A, Muniz RP
and Esquivel DM: beta-Lapachone enhancement of lipid peroxidation
and superoxide anion and hydrogen peroxide formation by sarcoma 180
ascites tumor cells. Biochem Pharmacol. 28:723–728. 1979.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Siegel D, Yan C and Ross D: NAD(P)H:
Quinone oxidoreductase 1 (NQO1) in the sensitivity and resistance
to antitumor quinones. Biochem Pharmacol. 83:1033–1040. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bentle MS, Reinicke KE, Bey EA, Spitz DR
and Boothman DA: Calcium-dependent modulation of poly(ADP-ribose)
polymerase-1 alters cellular metabolism and DNA repair. J Biol
Chem. 281:33684–33696. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Davalli P, Mitic T, Caporali A, Lauriola A
and D'Arca D: ROS, cell senescence and novel molecular mechanisms
in aging and age-related diseases. Oxid Med Cell Longev.
2016:35651272016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schieber M and Chandel NS: ROS function in
redox signaling and oxidative stress. Curr Biol. 24:R453–R462.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Raj L, Ide T, Gurkar AU, Foley M, Schenone
M, Li X, Tolliday NJ, Golub TR, Carr SA, Shamji AF, et al:
Selective killing of cancer cells by a small molecule targeting the
stress response to ROS. Nature. 475:231–234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Itoh K, Chiba T, Takahashi S, Ishii T,
Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, et
al: An Nrf2/small Maf heterodimer mediates the induction of phase
II detoxifying enzyme genes through antioxidant response elements.
Biochem Biophys Res Commun. 236:313–322. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Taguchi K, Motohashi H and Yamamoto M:
Molecular mechanisms of the Keap1-Nrf2 pathway in stress response
and cancer evolution. Genes Cells. 16:123–140. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kasai S, Arakawa N, Okubo A, Shigeeda W,
Yasuhira S, Masuda T, Akasaka T, Shibazaki M and Maesawa C:
NAD(P)H: Quinone oxidoreductase-1 expression sensitizes malignant
melanoma cells to the HSP90 inhibitor 17-AAG. PLoS One.
11:e01531812016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Miura S, Shibazaki M, Kasai S, Yasuhira S,
Watanabe A, Inoue T, Kageshita Y, Tsunoda K, Takahashi K, Akasaka
T, et al: A somatic mutation of the KEAP1 gene in malignant
melanoma is involved in aberrant NRF2 activation and an increase in
intrinsic drug resistance. J Invest Dermatol. 134:553–556. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Satoh T, Kosaka K, Itoh K, Kobayashi A,
Yamamoto M, Shimojo Y, Kitajima C, Cui J, Kamins J, Okamoto S, et
al: Carnosic acid, a catechol-type electrophilic compound, protects
neurons both in vitro and in vivo through activation of the
Keap1/Nrf2 pathway via S-alkylation of targeted cysteines on Keap1.
J Neurochem. 104:1116–1131. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schindelin J, Arganda-Carreras I, Frise E,
Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S,
Schmid B, et al: Fiji: An open-source platform for biological-image
analysis. Nat Methods. 9:676–682. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Okubo A, Yasuhira S, Shibazaki M,
Takahashi K, Akasaka T, Masuda T and Maesawa C: NAD(P)H
dehydrogenase, quinone 1 (NQO1), protects melanin-producing cells
from cytotoxicity of rhododendrol. Pigment Cell Melanoma Res.
29:309–316. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bang W, Jeon YJ, Cho JH, Lee RH, Park SM,
Shin JC, Choi NJ, Choi YH, Cho JJ, Seo JM, et al: β-lapachone
suppresses the proliferation of human malignant melanoma cells by
targeting specificity protein 1. Oncol Rep. 35:1109–1116. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nishimura K, Fukagawa T, Takisawa H,
Kakimoto T and Kanemaki M: An auxin-based degron system for the
rapid depletion of proteins in nonplant cells. Nat Methods.
6:917–922. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Venugopal R and Jaiswal AK: Nrf1 and Nrf2
positively and c-Fos and Fra1 negatively regulate the human
antioxidant response element-mediated expression of NAD(P)H:
Quinone oxidoreductase1 gene. Proc Natl Acad Sci USA.
93:14960–14965. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Singh A, Misra V, Thimmulappa RK, Lee H,
Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E,
et al: Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung
cancer. PLoS Med. 3:e4202006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ferlay J, Steliarova-Foucher E,
Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D and
Bray F: Cancer incidence and mortality patterns in Europe:
Estimates for 40 countries in 2012. Eur J Cancer. 49:1374–1403.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Marks R: Epidemiology of melanoma. Clin
Exp Dermatol. 25:459–463. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jackson JK, Higo T, Hunter WL and Burt HM:
Topoisomerase inhibitors as anti-arthritic agents. Inflamm Res.
57:126–134. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li CJ, Averboukh L and Pardee AB:
Beta-Lapachone, a novel DNA topoisomerase I inhibitor with a mode
of action different from camptothecin. J Biol Chem.
268:22463–22468. 1993.PubMed/NCBI
|
|
34
|
Li CJ, Li YZ, Pinto AV and Pardee AB:
Potent inhibition of tumor survival in vivo by beta-lapachone plus
taxol: Combining drugs imposes different artificial checkpoints.
Proc Natl Acad Sci USA. 96:13369–13374. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ough M, Lewis A, Bey EA, Gao J, Ritchie
JM, Bornmann W, Boothman DA, Oberley LW and Cullen JJ: Efficacy of
beta-lapachone in pancreatic cancer treatment: Exploiting the
novel, therapeutic target NQO1. Cancer Biol Ther. 4:95–102. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chakrabarti G, Silvers MA, Ilcheva M, Liu
Y, Moore ZR, Luo X, Gao J, Anderson G, Liu L, Sarode V, et al:
Tumor-selective use of DNA base excision repair inhibition in
pancreatic cancer using the NQO1 bioactivatable drug, β-lapachone.
Sci Rep. 5:170662015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yamaguchi Y, Hearing VJ, Maeda A and
Morita A: NADPH:quinone oxidoreductase-1 as a new regulatory enzyme
that increases melanin synthesis. J Invest Dermatol. 130:645–647.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kosaka K and Yokoi T: Carnosic acid, a
component of rosemary (Rosmarinus officinalis L.), promotes
synthesis of nerve growth factor in T98G human glioblastoma cells.
Biol Pharm Bull. 26:1620–1622. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Martin D, Rojo AI, Salinas M, Diaz R,
Gallardo G, Alam J, De Galarreta CM and Cuadrado A: Regulation of
heme oxygenase-1 expression through the phosphatidylinositol
3-kinase/Akt pathway and the Nrf2 transcription factor in response
to the antioxidant phytochemical carnosol. J Biol Chem.
279:8919–8929. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rau O, Wurglics M, Paulke A, Zitzkowski J,
Meindl N, Bock A, Dingermann T, Abdel-Tawab M and
Schubert-Zsilavecz M: Carnosic acid and carnosol, phenolic
diterpene compounds of the labiate herbs rosemary and sage, are
activators of the human peroxisome proliferator-activated receptor
gamma. Planta Med. 72:881–887. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Visanji JM, Thompson DG and Padfield PJ:
Induction of G2/M phase cell cycle arrest by carnosol and carnosic
acid is associated with alteration of cyclin A and cyclin B1
levels. Cancer Lett. 237:130–136. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Aruoma OI, Halliwell B, Aeschbach R and
Loligers J: Antioxidant and pro-oxidant properties of active
rosemary constituents: Carnosol and carnosic acid. Xenobiotica.
22:257–268. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Park SY, Song H, Sung MK, Kang YH, Lee KW
and Park JH: Carnosic acid inhibits the epithelial-mesenchymal
transition in B16F10 melanoma cells: A possible mechanism for the
inhibition of cell migration. Int J Mol Sci. 15:12698–12713. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Barni MV, Carlini MJ, Cafferata EG,
Puricelli L and Moreno S: Carnosic acid inhibits the proliferation
and migration capacity of human colorectal cancer cells. Oncol Rep.
27:1041–1048. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang X, Motea EA, Moore ZR, Yao J, Dong
Y, Chakrabarti G, Kilgore JA, Silvers MA, Patidar PL, Cholka A, et
al: Leveraging an NQO1 bioactivatable drug for tumor-selective use
of poly (ADP-ribose) polymerase inhibitors. Cancer Cell.
30:940–952. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
1000 Genomes Project Consortium, .
Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker
RE, Kang HM, Marth GT and McVean GA: An integrated map of genetic
variation from 1,092 human genomes. Nature. 491:56–65. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Fagerholm R, Hofstetter B, Tommiska J,
Aaltonen K, Vrtel R, Syrjakoski K, Kallioniemi A, Kilpivaara O,
Mannermaa A, Kosma VM, et al: NAD(P)H:quinone oxidoreductase 1
NQO1*2 genotype (P187S) is a strong prognostic and predictive
factor in breast cancer. Nat Genet. 40:844–853. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jamieson D, Cresti N, Bray J, Sludden J,
Griffin MJ, Hawsawi NM, Famie E, Mould EV, Verrill MW, May FE and
Boddy AV: Two minor NQO1 and NQO2 alleles predict poor response of
breast cancer patients to adjuvant doxorubicin and cyclophosphamide
therapy. Pharmacogenet Genomics. 21:808–819. 2011. View Article : Google Scholar : PubMed/NCBI
|