Introduction
Lung cancer, being the leading cause of
cancer-related mortality, results in over 1 million deaths per year
worldwide. Non-small cell lung cancer (NSCLC) accounts for about
85% of lung cancer cases. The prognosis of NSCLC is poor, with a
5-year overall survival rate of less than 20% (1). Εpidermal growth factor receptor
(EGFR) is a transmembrane receptor tyrosine kinase
frequently over-expressed in NSCLC (2). Activation of EGFR induces the
proliferation and growth of cancer cells and thus EGFR is a
promising target for personalized therapy. Recently, EGFR-tyrosine
kinase inhibitor (TKI) such as Gefitinib (AstraZeneca, London, UK),
Erlotinib (Genentech, San Francisco, USA) and Afatinib (Boehringer
Ingelheim, Ingelheim, Germany) have emerged as the first-line
therapy for the treatment of NSCLC (3–5). Through
selectively binding at the ATP-binding site of the receptor kinase
domain, TKIs block EGFR activation and its downstream signal
transduction, such as STAT3, MAP kinase (MAPK) and AKT signaling
(6). Although the EGFR-TKIs show a
rapid and dramatic tumor regression in NSCLC patients, the initial
clinical study with Gefitinib in 2005 illustrated that only 9–19%
of unselected patients with advanced NSCLC showed responses to the
treatment (7,8). Subsequently, researchers reported that
EGFR mutation status may be a key predictive factor in
determining the efficacy of EGFR-TKIs treatment (9). Exon 19 deletions (19Del) and
L858R substitution mutations were found to be the most
common EGFR mutations, which constitute about 90% of all
EGFR mutations (10). Several
studies revealed that NSCLC patients with EGFR mutations
showed better prognosis on EGFR-TKIs therapy. This was validated by
two phase II clinical trials, which demonstrated an overall
response rate of 75–78% for Gefitinib in NSCLC patients harboring
EGFR mutations (11,12). Nevertheless, secondary mutations
frequently arise after first-generation EGFR-TKIs treatment and
lead to therapeutic resistance. T790M mutation is the major
cause of resistance to first-generation EGFR-TKIs, accounting for
about 70% of acquired resistance (13). T790M mutation was reported to
associate with a shorter progression-free survival (PFS) with
EGFR-TKIs therapy in NSCLC patients (14). The relative EGFR mutant
abundance may also be a key factor in predicting the sensitivity of
EGFR-TKIs. Zhou et al reported that advanced NSCLC patients
with a high abundance of EGFR mutations showed a significant
longer PFS after Gefitinib treatment (15). In another study, Zhao et al
demonstrated that the abundance of EGFR mutations could be a
possible predictor to select patients with acquired resistance to
primary TKIs that would benefit from EGFR-TKIs re-administration
(16). Since the afore-mentioned
EGFR mutations are critical for deciding the treatment
strategies in patients with NSCLC, a reliable method to identify
and measure the EGFR mutant abundance is urgently
needed.
In the past decade, direct DNA sequencing was the
most frequently used method for EGFR mutation detection.
Despite its accessibility, the sensitivity is relatively low. The
detection limit of direct DNA sequencing is approximately 20%,
which may lead to a high rate of false negative result (17). More importantly, performing direct DNA
sequencing requires a long processing time and could delay
clinical-decision making. Currently, numerous detection methods
have been investigating for the detection of EGFR mutations
with a higher sensitivity (18,19).
Amplification refractory mutation system (ARMS) is a detection
method based on allele-specific polymerase chain reaction (PCR).
The detection limit is reported to be around 1% which is far more
sensitive than direct DNA sequencing. Droplet digital PCR (ddPCR)
is another emerging method to detect EGFR mutations. Based
on the compartmentalization and amplification of single DNA
molecules, ddPCR is highly sensitive. Thus, it is now generally
accepted as the most sensitive method for the detection and
quantification of mutations (20).
However, this method requires specialized equipment and it is more
expensive in terms of cost and labor. Here, we introduce a modified
ARMS-based assay, namely ARMS-Plus, for the analysis of EGFR
mutations based on real time PCR platform. In the present study,
ARMS-Plus and ddPCR methods were used to analyze FFPE-derived DNA
samples, with the aim of comparing the performances of the two
methods and exploring the feasibility of using ARMS-Plus in the
measuring of EGFR mutant abundance.
Materials and methods
Sample collection and processing
FFPE tissue samples obtained from patients between
2013 and 2015 were collected from the Guangdong Provincial Hospital
of Traditional Chinese Medicine. Written informed consent was
obtained from all of the patients. The present study protocol was
approved by the Institutional Review Board of the Guangdong
Provincial Hospital of Traditional Chinese Medicine. The FFPE
tissue samples were micro-dissected by a pathologist and assessed
by hematoxylin and eosin staining. Genomic DNA was extracted using
the QIAamp® DNA FFPE Tissue kit (Qiagen, Shanghai,
China). Collected FFPE-derived DNA samples were then analyzed by
the following assays.
ARMS-plus
The isolated DNA samples were processed by Human
EGFR Gene Mutation Quantitative Detection kit (Fluorescenece
qPCR; GenoSaber Biotech Co., Ltd., Shanghai, China) according to
manufacturer's instructions. This is a quantitative PCR kit that
can detect 4 different EGFR mutations with an external
control in exon 4 to monitor samples qualities (Table I). In brief, 5 µl samples were added
to the pre-mixed 45 µl reaction mixture came with the kit and the
PCR was conducted with Applied Biosystems® 7500
Real-Time PCR Systems (Thermo Fisher Scientific, Inc., Waltham, MA,
USA). PCR amplification conditions were as follow: hot start at
95°C for 4 min, and 50 cycles of 95°C for 10 sec, 61°C for 30 sec
with fluorescence reading (FAM). Primer and probe sequences were
listed in Table II. 19Del
mutation was detected by two separate reactions as two different
kinds of base pair alteration were found. We also found that the
2361-site single nucleotide polymorphism (SNP) may affect PCR
specificity, hence the T790M test was divided into two
reactions to raise detection rate. The mutant abundance was defined
as the percentage of mutant copies present in the whole DNA sample.
To calculate the copy number, serially diluted plasmids were
provided in the kit as standard samples for generating the
standardization curve. Theoretically, the calculation equation of
mutant abundance is shown as follow:
 | Table I.Mutations or gene fragments detected
by ARMS-Plus. |
Table I.
Mutations or gene fragments detected
by ARMS-Plus.
| Site | Mutation | Exon | Base pair
altered | Cosmic ID |
|---|
| EGFR
external control | / | 4 | / | / |
| Del(1) |
E746_A750del(1) | 19 | 2235_2249del15 | 6223 |
| Del(2) |
E746_A750del(2) | 19 | 2236_2250del15 | 6225 |
| L858R | L858R | 21 | 2573T>G | 6224 |
| T790M(1) | T790M | 20 | 2369C>T | 6240 |
| T790M(2) | T790M | 20 |
2361G>A/2369C>T | 1451600/6240 |
 | Table II.Sequence of primers and probes
utilized for ARMS-Plus. |
Table II.
Sequence of primers and probes
utilized for ARMS-Plus.
| Site | Primer | Probe | Blocker |
|---|
| EGFR external
control | Forward:
5′-TGGAGAGCATCCAGT-3′ |
5′FAM-ACATAGTCAGCAGTGACTT-3′MGB | / |
|
| Reverse:
5′-TCTGGAAGTCCATCGACAT-3′ |
|
|
| Del(1) | Forward:
5′-CCGTCGCTATCAAA-3′ |
5′FAM-AAGCCAACAAGGAAAT-3′MGB | / |
|
| Reverse:
5′-ATGGACCCCCACAC-3′ |
|
|
| Del(2) | Forward:
5′-GTCGCTATCAAGA-3′ |
5′FAM-AAGCCAACAAGGAAAT-3′MGB | / |
|
| | |
|
| Reverse:
5′-GTCGCTATCAAGA-3′ |
|
|
| L858R | Forward:
5′-AGATTTTGGGCG-3′ |
5′FAM-AACTGCTGGGTGCGGA-3′MGB |
5′-TTTTTGGGCGGGCCAAAC-3′
phosphorthioated |
|
| Reverse:
5′-TTTGCCTCCTTCTGC-3′ |
|
|
| T790M(1) | Forward:
5′-CCGTGCAGCACATCAT-3′ |
5′FAM-CTCATGCCCTTCGGC-3′MGB |
5′-GCTCATCACGCAGCTCA-3′
phosphorthioated |
|
| Reverse:
5′-TGTCTTTGTGTTCCCG-3′ |
|
|
| T790M(2) | Forward:
5′-CCGTGCAACACATCAT-3′ |
5′FAM-CTCATGCCCTTCGGC-3′MGB |
5′-CAACTCATCACGCAGCT-3′
phosphorthioated |
|
| Reverse:
5′-TGTCTTTGTGTTCCCG-3′ |
|
|
Mutant abundance=Observed mutant copy
numberObserved wild type copy number
ddPCR
ddPCR was performed as previously described
(21). In brief, the mixture of
probes, primers, template DNA and 2X Droplet PCR Supermix (Bio-Rad
Laboratories, Hercules, CA, USA) was applied to the droplet
generator. The oil-in-water mixture generated was then transferred
to a 96-well PCR plate, heat-sealed and placed in a C1000 Touch
thermal cycler (Bio-Rad Laboratories) for PCR amplification. The
following thermo-cycling conditions were used: Hot start at 95°C
for 10 min, 40 cycles of 94°C for 30 sec, 62°C for 1 min, and 98°C
for 10 min. The PCR product was read on a QX-200 droplet reader
(Bio-Rad Laboratories) and the data were analyzed with Quantilife
software. The Qunatilife software calculates the copy number of
both mutant and wild-type DNA based on the Poisson statistic.
ARMS-PCR
The isolated DNA samples were re-tested with ADx
EGFR Mutations Detection kit (Amoy Diagnostics, Xiamen, China),
which has received China Food and Drug Administration's (CFDA)
approval for clinical usage in mainland China. The kit covered the
29 EGFR mutation hotspots from exon 18 to 21. The assay was
carried out according to the manufacturer's protocol with the
MX3000P (Stratagene; Agilent Technologies, Inc., Santa Clara, CA,
USA) Real-Time PCR system.
Statistical analysis
The concordance rate of ARMS-Plus and ddPCR in
identifying EGFR mutation status was analyzed by Kappa test.
Samples were examined to determine whether a statistical difference
existed regarding variations in EGFR mutations between
ARMS-Plus and ddPCR by the Wilcoxon test. P<0.05 was considered
to indicate statistical significance. The correlation between
ARMS-Plus and ddPCR results on the EGFR mutant abundance was
examined by Linear Regression analysis. The statistical analysis
and graphic generation were carried out by using SAS9.2 (SAS
Institute, Inc., Cary, NC, USA) and GraphPad Prism 5 (GraphPad
Software, Inc., La Jolla, CA, USA).
Results
Evaluation of the detection efficiency
of ARMS-Plus
Mutant cell lines H1650 (19Del) and H1975
(L858R and T790M) were spiked into EGFR
wild-type cell line A549 at a ratio of 1 and 5% (2×103
and 1×104 cells of each H1650 and H1975 was spiked into
2×105 cells of A549, respectively). QIAamp DNA FFPE
Tissue Kit (Qiagen) was used to extract DNA from the cell mixtures.
The final allelic frequencies were analyzed by both ARMS-Plus and
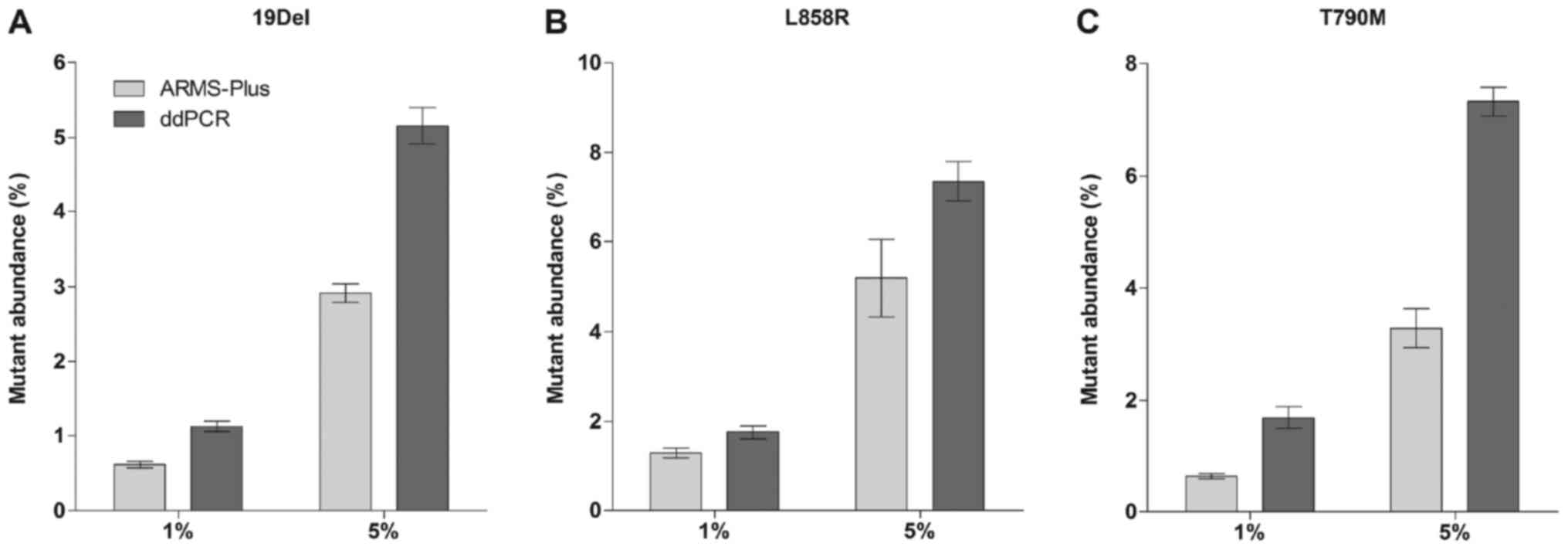
ddPCR (3 repeats for each cell mixture). Results demonstrated that
all mutations were stably detected by ARMS-Plus (Fig. 1). Generally, the mutant abundance
detected by ddPCR was higher than that by ARMS-Plus.
Patients' enrollment
FFPE tumor samples from 77 lung adenocarcinoma
patients were collected in this study (Table III). Of these patients, 45% (35/77)
were female, median age was 62 years (range from 37–91 years), and
52% were never-smokers. Nearly half of the patients (45%, 35/77)
were diagnosed with advanced stage disease (stage III–IV). Only 4
patients were treated with EGFR-TKI before sample collection.
 | Table III.Patient characteristics. |
Table III.
Patient characteristics.
|
|
| EGFR
mutation status detected by ddPCR |
|---|
|
|
|
|
|---|
| Variable | n (%) | Mutant | Wild-type |
|---|
| Sex |
|
|
|
|
Female | 35 (45) | 24 | 11 |
|
Male | 42 (55) | 11 | 31 |
| Age |
|
|
|
| ≥60
years | 46 (60) | 15 | 31 |
| <60
years | 31 (40) | 20 | 11 |
| Smoking
history |
|
|
|
|
Ever-smoker | 33 (43) | 9 | 24 |
|
Never-smoker | 40 (52) | 26 | 14 |
|
Uncertain | 4 (5) | 0 | 4 |
| Disease stage |
|
|
|
| I | 7 (9) | 5 | 2 |
| II | 26 (34) | 12 | 14 |
|
III | 10 (13) | 5 | 5 |
| IV | 25 (32) | 12 | 13 |
|
Uncertain | 9
(12) | 1 | 8 |
| EGFR mutation
type |
|
|
|
|
19Del | 21 (27) | 21 | 0 |
|
L858R | 12 (16) | 12 | 0 |
|
19Del+T790M | 2 (3) | 2 | 0 |
|
Wild-type | 42 (54) | 0 | 42 |
| Histopathology
type |
|
|
|
|
Adenocarcinoma | 77 (100) | 35 | 42 |
| Total
n | 77 | 35 | 42 |
Determination of EGFR mutation
status
ARMS-Plus was performed to analyze the EGFR
mutation status. The results were then re-tested by ddPCR and
ARMS-PCR (Table IV).
 | Table IV.EGFR mutation status detected
by ARMS-PCR, ddPCR and ARMS-Plus. |
Table IV.
EGFR mutation status detected
by ARMS-PCR, ddPCR and ARMS-Plus.
| Mutation | ARMS-PCR | ddPCR | ARMS-Plus |
|---|
| Wild type | 43 | 42 | 41 |
| 19Del | 20 | 21 | 21 |
| L858R | 11 | 12 | 13 |
|
19Del+L858R | 1 | 0 | 0 |
|
19Del+T790M | 2 | 2 | 2 |
| Total n | 77 | 77 | 77 |
Among the 77 paired samples evaluated by ddPCR, 35
(45.5%) samples were mutation positive and the other 42 samples
were negative. For the ARMS-Plus results, only one sample showed a
discordant result compared to that of ddPCR. A case of weak
positive L858R result with an abundance of 0.5% detected by
ARMS-Plus was recognized as negative by ddPCR (Table V). Together, the overall concordance
rate of EGFR mutation testing between ARMS-Plus and ddPCR
was 98.7% (76/77). The results of EGFR mutation status
identified by the two methods were highly consistent (Kappa=0.9739,
95% CI: 0.923–1).
 | Table V.Comparison of EGFR mutation
status detected by ARMS-Plus and ddPCR. |
Table V.
Comparison of EGFR mutation
status detected by ARMS-Plus and ddPCR.
|
| ARMS-Plus
results |
|
|---|
|
|
|
|
|---|
| Variable | EGFR Mutant
(n) | Wild-type (n) | Total |
|---|
| ddPCR results |
|
|
|
| EGFR
Mutant (n) | 35 | 0 | 35 |
|
Wild-type (n) | 1 | 41 | 42 |
|
Total | 36 | 41 | 77 |
To evaluate the utility of ARMS-Plus for identifying
EGFR mutations, we assessed the sensitivity and specificity
of the assay compared to ddPCR (Table
VI). For 19Del and T790M mutations, ARMS-Plus
results were 100% matched with that of ddPCR. Only one case of
L858R mutation was detected in ARMS-Plus but not ddPCR. The
overall sensitivity and specificity for detecting EGFR
mutations were 100 and 98% respectively, demonstrating the promise
of ARMS-Plus in identifying EGFR mutations.
 | Table VI.Performance of ARMS-Plus compared
with ddPCR. |
Table VI.
Performance of ARMS-Plus compared
with ddPCR.
| Mutation | Sensitivity
(%) | Specificity
(%) | PPV (%) | NPV (%) | Total n number |
|---|
| 19Del | 100 | 100 | 100 | 100 | 23 |
| L858R | 100 | 98 | 92 | 100 | 13 |
| T790M | 100 | 100 | 100 | 100 | 2 |
| Total | 100 | 98 | 97 | 100 | 36 |
The ARMS-PCR results were also similar to that of
ARMS-Plus and ddPCR. Only one case of positive 19Del result
confirmed by both ARMS-Plus and ddPCR was classified as negative by
ARMS-PCR and one case of negative 19Del result detected by
both ARMS-Plus and ddPCR was shown positive for ARMS-PCR.
Collectively, these data indicate that both ARMS-PCR, ddPCR and
ARMS-Plus results were consistent in identifying EGFR
mutations in FFPE-derived DNA samples.
Quantitative analysis of EGFR
mutations
23 out of 77 samples were identified to be
19Del mutant by both ARMS-Plus and ddPCR. According to the
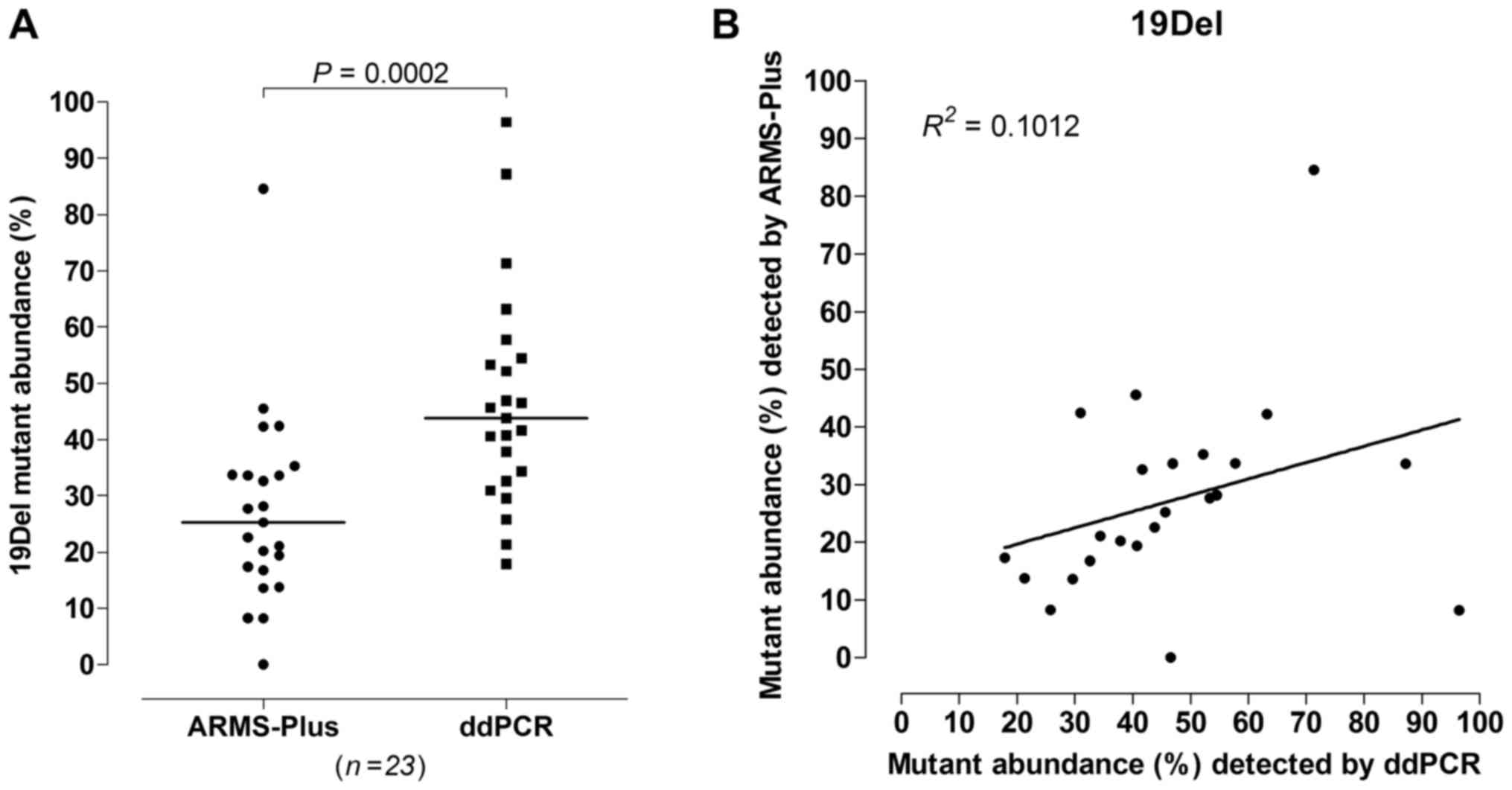
results, significant difference was observed (Wilcoxon rank sum
test, P=0.0002, Fig. 2A) between the
abundance of 19Del mutation detected by ARMS-Plus (median=25.26%)
and ddPCR (median=43.76%). The observed 19Del mutant
abundance detected by ARMS-Plus was not consistent with the
expected abundance based on ddPCR (Linear regression analysis,
R2=0.1012, Fig.
2B).
For L858R mutation, 12 out of 77 samples were
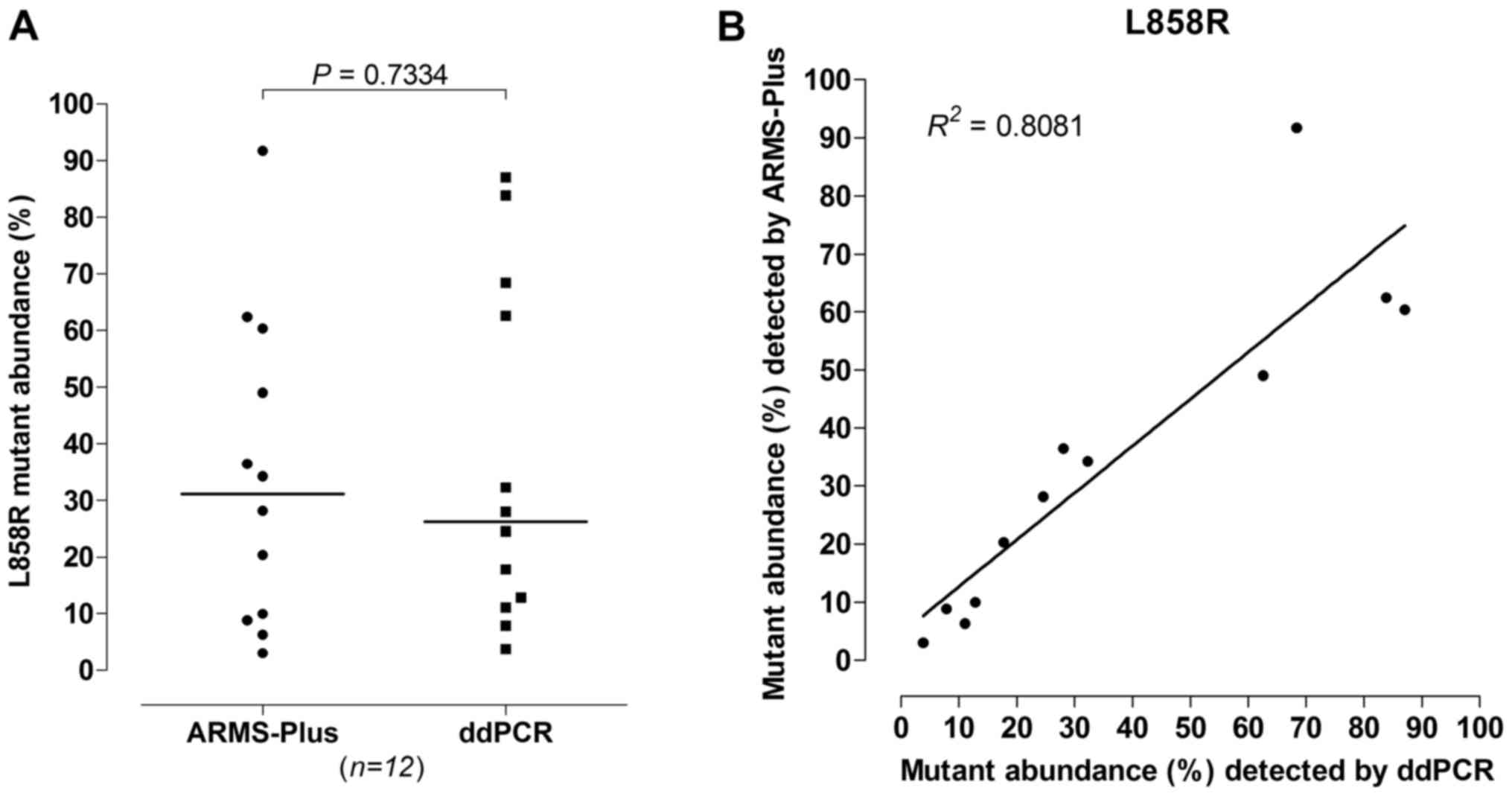
identified by both ARMS-Plus and ddPCR. No significant difference
was observed (Wilcoxon rank sum test, P=0.7334, Fig. 3A) between the abundance of
L858R mutation detected by ARMS-Plus (median=31.18%) and
ddPCR (median=26.27%). The observed L858R mutant abundance
detected by ARMS-Plus was highly consistent with the expected
abundance based on ddPCR (Linear regression analysis,
R2=0.8081, Fig.
3B).
The data of T790M mutant abundance were not
analyzed as only 2 samples were identified with T790M
mutation (both patients were EGFR-TKI treatment-naïve). However,
the observed mutant abundance detected by ARMS-Plus is generally
consistent with that of ddPCR (Case 1: 29.21% and 43.17%; Case 2:
11.18 and 16.42%, for ARMS-Plus and ddPCR, respectively).
Discussion
Since the approval of EGFR-TKIs in the last decade,
a massive amount of researches have been investigating the clinical
significance of EGFR mutations in predicting the sensitivity
of TKIs treatment (22,23). It is now apparent that EGFR
mutations are associated with the efficacy of TKIs therapy. All
NSCLC patients are suggested to perform EGFR mutation test
after diagnosis to save cost and avoid unnecessary adverse effects
due to wrong regimen decision (24).
Recently, clinical study revealed that Osimertinib, a
third-generation EGFR-TKI that can overcome T790M-resistance,
showed a superior efficacy as first-line treatment to
EGFR-mutated advanced NSCLC compared to first-generation
EGFR-TKIs (25). Hence, a sensitive
assay that can identify patients with ‘de novo’ T790M
mutation could be valuable for selecting patients who are favorable
to first-line Osimertinib treatment.
As NSCLC are often detected at advanced stages,
small diagnostic biopsy specimens are the primary source for
mutation detection. The DNA amount in these specimens is usually
limited due to prior morphologic and immunophenotypic analysis.
Thus, a highly sensitive assay is required for the detection of
EGFR mutations. ddPCR is now generally accepted as the most
sensitive method for evaluating mutations with low mutation rate.
It utilizes a water-oil emulsion droplet system to separate DNA
samples into thousands of nanoliter-sized droplets. The
partitioning enables the measurement of thousands of independent
amplification events within a single sample and extensively raises
the sensitivity of the assay. In 2014, Zhang et al reported
the superiority of ddPCR in detecting EGFR mutations.
Results illustrated that ddPCR was able to detect and quantify
EGFR mutations for as low as 0.12–2.73% (26). Despite the high sensitivity of ddPCR,
the high cost and the requirement of unique equipment have greatly
hampered its clinical application.
In the present study, we interpreted the ability of
ARMS-Plus in identifying and quantifying EGFR mutations,
with ddPCR as a standard. ARMS-Plus is an AMRS-based assay
optimized for the quantification of 4 different EGFR
mutations, including 19Del at two different sites,
L858R, and T790M mutations. Although only limited
mutation sites are analyzed by ARMS-Plus, they represent the
majority of mutations which have a well-established clinical
significance in guiding EGFR-TKIs treatment selection. Moreover,
the primers and probes of ARMS-Plus are optimized for EGFR
mutation detection and the amplicon of each quantitative PCR is
shortened to 50–80 bp. These features enable the precise analysis
of highly fragmented DNA molecules extracted from FFPE tumor
samples. To specifically quantify EGFR mutations, a kind of
3′end-phosphorylated blocking primer was employed into the
ARMS-Plus PCR reaction pools. By complementing the wild type DNA at
the mutated sites, these primers block the non-specific elongations
and raise the accuracy in quantification. In addition, ARMS-Plus
can be conveniently performed with a real-time PCR system. These
features of ARMS-Plus allow a rapid, reliable and cost-effective
molecular analysis in clinical practice.
Cell experiment was performed to evaluate the
detection efficiency of ARMS-Plus. EGFR mutations from
well-studied cell lines (H1650 and H1975) were stably detected by
both ARMS-Plus and ddPCR assays. Generally, the mutant abundance
detected by ddPCR were higher than that by ARMS-Plus. This could be
due to the possible non-specific amplification of wild-type
template in ddPCR.
To explore the sensitivity of ARMS-Plus, a total of
77 FFPE tumor samples were analyzed for EGFR mutation
statuses by both ARMS-PCR, ddPCR and ARMS-Plus. Micro-dissection
was performed by a pathologist before the tests to remove normal
tissues presented in the FFPE samples which could affect the
quantification of mutations. All mutations found by ddPCR (35/77)
were consistent with the results of ARMS-Plus. Only one case of
weak positive L858R mutation with an abundance of 0.5% was
identified by ARMS-Plus, but not by ddPCR and ARMS-PCR. The overall
concordance rate of EGFR mutation testing was 98.7%
(Kappa=0.9739). These results demonstrated that ARMS-Plus is a
promising assay with high sensitivity in detecting EGFR
mutations.
In the quantification of EGFR mutations, the
results varied with different types of mutation. For L858R
mutation, there was no significant difference (P=0.7334) between
the abundance detected by ARMS-Plus and ddPCR and a clear positive
correlation was observed (R2=0.8081). However, a
significant difference was found (P=0.0002) between the abundance
of 19Del mutation detected by ARMS-Plus and ddPCR. The
19Del mutant abundance detected by ARMS-Plus was not
consistent with the expected abundance based on ddPCR
(R2=0.1012). The duplex probes used in ddPCR for
detecting 19Del mutation cover a larger region than that in
ARMS-Plus (ddPCR: K749-S752 and A755-D761 region, ARMS-Plus:
E746_A750 region) (21). This could
probably explain the difference in 19Del mutant abundance
detected by the two methods, where the results of ddPCR were
significantly higher than that of ARMS-Plus (median=43.76% vs.
25.26%). Further research with a larger sample size is required to
clarify the ability of ARMS-Plus in evaluating 19Del mutant
abundance. For T790M mutation, because only 2 samples were
identified, no analysis is performed to evaluate its correlation
between ARMS-Plus and ddPCR.
There were a few limitations in this work. First,
FFPE samples can only represents a unique tumor cell clones and
cannot reflect the genetic heterogeneity of the whole tumor. The
high sensitivity molecular screening for detection of EGFR
mutations is at risk of over-qualification for EGFR-TKIs therapy.
Therefore, combining liquid biopsy is a better option for decision
making. Indeed, ddPCR has been proved to be sensitive enough to
detect EGFR mutations in plasma samples (27). Further study to investigate the
performance of ARMS-Plus in liquid biopsy is required. Second, due
to the small sample size of this study, the performance of
ARMS-Plus in quantifying T790M mutation cannot be assessed.
Third, although micro-dissection was performed to ensure a >80%
tumor content in each sample, the variation in tumor content could
cause inaccurate quantification.
In conclusion, ARMS-Plus provides a reliable and
cost-effective quantitative measuring tool with a similar
sensitivity and specificity to ddPCR and could be applicable for
evaluating EGFR mutations and guiding regimen selection in
lung adenocarcinoma patients.
Acknowledgements
The authors Dr ZW Guo and Dr GH Yang are employees
of GenoSaber Biotech Co., Ltd. (Shanghai, China). This company
provided the Human EGFR Gene Mutation Quantitative Detection
kit (Fluorescence qPCR) used in the present study and also provided
technical support.
Glossary
Abbreviations
Abbreviations:
|
ARMS
|
amplification refractory mutation
system
|
|
CFDA
|
China Food and Drug Administration
|
|
PCR
|
polymerase chain reaction
|
|
ddPCR
|
droplet digital PCR
|
|
EGFR
|
epidermal growth factor receptor
|
|
19Del
|
exon 19 deletion
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
MAPK
|
mitogen-activated protein kinase
|
|
NSCLC
|
non-small cell lung cancer
|
|
PFS
|
progression-free survival
|
|
SNP
|
single nucleotide polymorphism
|
|
TKIs
|
tyrosine kinase inhibitors
|
References
|
1
|
National Comprehensive Cancer Network
(NCCN), . Non-small cell lung cancer, Version 8.2017. NCCN Clinical
Practice Guidelines in Oncology. NCCN; Fort Washington, PA:
2017
|
|
2
|
Boch C, Kollmeier J, Roth A,
Stephan-Falkenau S, Misch D, Grüning W, Bauer TT and Mairinger T:
The frequency of EGFR and KRAS mutations in non-small cell lung
cancer (NSCLC): Routine screening data for central Europe from a
cohort study. BMJ Open. 3:pii: e0025602013. View Article : Google Scholar
|
|
3
|
Paez JG, Jänne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kayye FJ, Lindeman N, Boggon TJ,
et al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rosell R, Carcereny E, Gervais R,
Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R,
Pallares C, Sanchez JM, et al: Erlotinib versus standard
chemotherapy as first-line treatment for European patients with
advanced EGFR mutation-positive non-small-cell lung cancer
(EURTAC): A multicentre, open-label, randomised phase 3 trial.
Lancet Oncol. 13:239–246. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sequist LV, Yang JC, Yamamoto N, O'Byrne
K, Hirsh V, Mok T, Geater SL, Orlov S, Tsai CM, Boyer M, et al:
Phase III study of afatinib or cisplatin plus pemetrexed in
patients with metastastic lung adenocarcinoma with EGFR mutation. J
Clin Oncol. 31:3327–3334. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mitsudomi T and Yatabe Y: Epidermal growth
factor receptor in relation to tumor development: EGFR gene and
cancer. FEBS J. 277:301–308. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fukuoka M, Yano S, Giaccone G, Tamura T,
Nakagawa K, Douillard JY, Nishiwaki Y, Vansteenkiste J, Kudoh S,
Rischin D, et al: Multi-institutional randomized phase II trial of
gefitinib for previously treated patients with advanced
non-small-cell lung cancer (The IDEAL 1 Trial) [corrected]. J Clin
Oncol. 21:2237–2246. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kris MG, Natale RB, Herbst RS, Lynch TJ
Jr, Prager D, Belani CP, Schiller JH, Kelly K, Spiridonidis H,
Sandler A, et al: Efficacy of gefitinib, an inhibitor of the
epidermal growth factor receptor tyrosine kinase, in symptomatic
patients with non-small cell lung cancer: A randomized trial. JAMA.
290:2149–2158. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Usuda K, Sagawa M, Motono N, Ueno M,
Tanaka M, Machida Y, Matoba M, Taniguchi M, Tonami H, Ueda Y and
Sakuma T: Relationships between EGFR mutation status of lung cancer
and preoperative factors-are they predictive? Asian Pac J Cancer
Prev. 15:657–662. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kosaka T, Yatabe Y, Endoh H, Kuwano H,
Takahashi T and Mitsudomi T: Mutations of the epidermal growth
factor receptor gene in lung cancer: Biological and clinical
implications. Cancer Res. 64:8919–8923. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sutani A, Nagai Y, Udagawa K, Uchida Y,
Koyama N, Murayama Y, Tanaka T, Miyazawa H, Nagata M, Kanazawa M,
et al: Gefitinib for non-small-cell lung cancer patients with
epidermal growth factor receptor gene mutations screened by peptide
nucleic acid-locked nucleic acid PCR clamp. Br J Cancer.
95:1483–1489. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Inoue A, Suzuki T, Fukuhara T, Maemondo M,
Kimura Y, Morikawa N, Watanabe H, Saijo Y and Nukiwa T: Prospective
phase II study of gefitinib for chemotherapy-naive patients with
advanced non-small-cell lung cancer with epidermal growth factor
receptor gene mutations. J Clin Oncol. 24:3340–3346. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shih JY, Gow CH and Yang PC: EGFR mutation
conferring primary resistance to gefitinib in non-small-cell lung
cancer. N Engl J Med. 353:207–208. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Su KY, Chen HY, Li KC, Kuo ML, Yang JC,
Chan WK, Ho BC, Chang GC, Shih JY, Yu SL and Yang PC: Pretreatment
epidermal growth factor receptor (EGFR) T790M mutation predicts
shorter EGFR tyrosine kinase inhibitor response duration in
patients with non-small-cell lung cancer. J Clin Oncol. 30:433–440.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou Q, Zhang XC, Chen ZH, Yin XL, Yang
JJ, Xu CR, Yan HH, Chen HJ, Su J, Zhong WZ, et al: Relative
abundance of EGFR mutations predicts benefit from gefitinib
treatment for advanced non-small-cell lung cancer. J Clin Oncol.
29:3316–3321. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao ZR, Wang JF, Lin YB, Wang F, Fu S,
Zhang SL, Su XD, Jiang L, Zhang YG, Shao JY and Long H: Mutation
abundance affects the efficacy of EGFR tyrosine kinase inhibitor
readministration in non-small-cell lung cancer with acquired
resistance. Med Oncol. 31:8102014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ogino S, Kawasaki T, Brahmandam M, Yan L,
Cantor M, Namgyal C, Mino-Kenudson M, Lauwers GY, Loda M and Fuchs
CS: Sensitive sequencing method for KRAS mutation detection by
Pyrosequencing. J Mol Diagn. 7:413–421. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cushman-Vokoun AM, Crowley AM, Rapp SA and
Greiner TC: Comparison study of the performance of the QIAGEN EGFR
RGQ and EGFR pyro assays for mutation analysis in non-small cell
lung cancer. Am J Clin Pathol. 140:7–19. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shaozhang Z, Ming Z, Haiyan P, Aiping Z,
Qitao Y and Xiangqun S: Comparison of ARMS and direct sequencing
for detection of EGFR mutation and prediction of EGFR-TKI efficacy
between surgery and biopsy tumor tissues in NSCLC patients. Med
Oncol. 31:9262014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ellison G, Donald E, McWalter G, Knight L,
Fletcher L, Sherwood J, Cantarini M, Orr M and Speake G: A
comparison of ARMS and DNA sequencing for mutation analysis in
clinical biopsy samples. J Exp Clin Cancer Res. 29:1322010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu Q, Zhu Y, Bai Y, Wei X, Zheng X, Mao M
and Zheng G: Detection of epidermal growth factor receptor mutation
in lung cancer by droplet digital polymerase chain reaction. Onco
Targets Ther. 8:1533–1541. 2015.PubMed/NCBI
|
|
22
|
Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong
V, Sandoval-Tan J, Ladrera G, Thongprasert S, Srimuninnimit V, Liao
M, et al: Detection and dynamic changes of EGFR mutations from
circulating tumor DNA as a predictor of survival outcomes in NSCLC
patients treated with first-line intercalated Erlotinib and
chemotherapy. Clin Cancer Res. 21:3196–3203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tseng JS, Yang TY, Tsai CR, Chen KC, Hsu
KH, Tsai MH, Yu SL, Su KY, Chen JJ and Chang GC: Dynamic plasma
EGFR mutation status as a predictor of EGFR-TKI efficacy in
patients with EGFR-mutant lung adenocarcinoma. J Thorac Oncol.
10:603–610. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lilenbaum RA and Horn LA: Management of
EGFR mutation-positive non-small cell lung cancer. J Natl Compr
Canc Netw. 14(5 Suppl): S672–S674. 2016. View Article : Google Scholar
|
|
25
|
Ramalingam SS, Yang JC, Lee CK, Kurata T,
Kim DW, John T, Nogami N, Ohe Y, Mann H, Rukazenkov Y, et al:
Osimertinib as first-line tratment of EGFR mutation-positive
advanced non-small-cell lung cancer. J Clin Oncol. Aug
25–2017.(Epub ahead of print). View Article : Google Scholar
|
|
26
|
Zhang BO, Xu CW, Shao Y, Wang HT, Wu YF,
Song YY, Li XB, Zhang Z, Wang WJ, Li LQ and Cai CL: Comparison of
droplet digital PCR and conventional quantitative PCR for measuring
EGFR gene mutation. Exp Ther Med. 9:1383–1388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu G, Ye X, Dong Z, Lu YC, Sun Y, Liu Y,
McCormack R, Gu Y and Liu X: Highly sensitive droplet digital PCR
method for detection of EGFR-activating mutations in plasma
cell-free DNA from patients with advanced non-small cell lung
cancer. J Mol Diagn. 17:265–272. 2015. View Article : Google Scholar : PubMed/NCBI
|