Introduction
Lung cancer is among the most common types of
cancer, accounting for ~13% of all cancer cases (1). The generally poor prognosis of lung
cancer renders it a leading cause of cancer-associated mortality
worldwide (2). In 2010, 1.5 million
mortalities due to lung cancer were reported, representing 19% of
all cancer-associated mortality (3).
The incidence of lung cancer has doubled in China over the past
decade due to issues including the aging population, smoking and
the reduced air quality (4). Lung
cancer is initiated by the activation of oncogenes or the
inactivation of tumor suppressor genes (5). Despite advances in diagnosis and
treatment, the prognosis of lung cancer remains relatively poor.
The identification of reliable biomarkers and novel genes involved
in lung cancer carcinogenesis is important for improving the
ability to predict the prognosis and to guide the therapy of lung
cancer.
Forkhead box D3 (FOXD3) is a member of the
FOX transcription factor family, which is characterized by a
distinct forkhead domain (6).
FOXD3 acts as a transcriptional repressor or activator
(7). The abnormal expression of
FOXD3 has been reported to participate in tumor onset and
progression in non-small cell lung cancer tumor cells (8). Other studies have indicated tumor
suppressive activities for FOXD3, including the inhibition
of cell growth and invasion in various types of cancer, including
gastric cancer and melanoma (9,10). A
number of genes associated with tumorigenesis have been reported to
be targets of FOXD3. One study demonstrated that
FOXD3 regulated RND3 expression and migration
properties in melanoma cells (11).
Another reported that FOXD3 exhibited tumor suppressive activity
that affected the growth, aggressiveness and angiogenesis of
neuroblastoma through the transcriptional regulation of
NDRG1 (12). However, the role
of FOXD3 in lung cancer remains uncharacterized.
In this study, differentially expressed genes (DEGs)
and alternative splicing genes (ASGs) were identified in
FOXD3-knockout samples compared with normal samples.
Functional and pathway enrichment analyses of the DEGs and ASGs
were performed. A protein-protein interaction (PPI) network was
constructed based on the overlaps between the DEGs and ASGs. An
improved understanding of FOXD3 in regulating the process of
lung cancer was obtained, which may allow the development of novel
strategies for the diagnosis and therapy of lung cancer.
Materials and methods
Datasets
The gene expression profile GSE64513 was downloaded
from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database. The data
set contained the RNAseq data from 6 samples, including 3
FOXD3-knockout A549 lung cancer cell samples and 3 normal
A549 cell samples.
Screening of DEGs and ASGs
The data was first analyzed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc),
a java-based high-throughput data quality control software. Reads
with base quality scores <20 were discarded, and reads longer
than 30 bp were selected for further investigation. The remaining
reads were mapped to the GRCh37/hg19 genome based on the Tophat2
program (13). The number of reads
mapped to the exons of each gene was counted with the HTSeq-Count
tool (14) and regarded as the
expression profile of each gene. Differently expressed genes (DEGs)
in FOXD3 knockout lung cancer samples compared with normal
samples were identified using the edge R package (15) with the following thresholds: False
discovery rate <0.01 and |log (fold change)| >1. The
hierarchical clustering of DEGs was performed using the heatmap.2
function of the gplots package in Various R Programming tool
(version 2.12) (16). The alternative
splicing genes (ASGs) in the FOXD3 knockout samples were
identified using the replicate multivariate analysis of transcript
splicing (rMATS) program, a computer program designed to detect
differential alternative splicing from replicate RNA-Seq data
(17).
Functional and pathway enrichment
analysis
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; https://david.ncifcrf.gov/) is a web-based tool for
genomic functional annotations (18).
To further explore the biological functions of the DEGs and ASGs,
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) enrichment analyses were performed using DAVID, with the
threshold of P<0.05.
Construction of a PPI network
The overlapping DEGs and ASGs were analyzed using
the Search Tool for the Retrieval of Interacting Genes (STRING;
http://string-db.org/) (19,20). A PPI
network to illustrate the identified interactions was constructed
and visualized using Cytoscape 3.4 (21).
Results
Identification of DEGs and ASGs
The total number of reads, the number of mapped
reads and the mapping rate of each sample is provided in Table I. A total of 1,853 DEGs were
identified, of which 382 were upregulated and 1,471 were
downregulated. The top 20 DEGs are listed in Table II. Fig.
1 demonstrates the hierarchical clustering results for each
sample graphically (Fig. 1A), the
fold-change trend of the expression of the identified DEGs
(Fig. 1B) and the hierarchical
cluster analysis of the samples based on the DEGs (Fig. 1C). A total of 2,249 genes with
alternative splicing were identified in FOXD3-knockout lung
cancer samples compared with normal A549 cell samples, including
545 with an alternative 3′ splice site, 412 with an alternative 5′
splice site, 1,629 with mutually exclusive exons and 67 with
retained introns.
 | Table I.Total reads, the number of mapped
reads, and the mapping rates of each sample. |
Table I.
Total reads, the number of mapped
reads, and the mapping rates of each sample.
| Sample | Total reads | Mapped reads | Mapping rate,
% |
|---|
| SRR1734826 | 10,339,232 | 9,252,784 | 89.5 |
| SRR1734827 | 10,472,212 | 9,298,751 | 88.8 |
| SRR1734828 | 10,868,010 | 9,651,798 | 88.8 |
| SRR1734829 | 11,224,483 | 9,666,662 | 86.1 |
| SRR1734830 | 10,548,877 | 9,241,415 | 87.6 |
| SRR1734831 | 11,578,464 | 10,104,885 | 87.3 |
 | Table II.Top 20 differentially expressed genes
of the forkhead Box D3-knockout lung cancer A549 cell samples
compared with normal A549 cells. |
Table II.
Top 20 differentially expressed genes
of the forkhead Box D3-knockout lung cancer A549 cell samples
compared with normal A549 cells.
| Gene symbol | False discovery
rate | P-value | Log fold
change |
|---|
| SAA1 |
1.02×10−426 |
8.46×10−313 | −7.70213 |
| C3 |
3.82×10−478 |
8.15×10−562 | −2.97374 |
| GAS6 |
9.26×10−283 |
2.01×10−286 | −2.52365 |
| CFB |
4.97×10−271 |
1.44×10−274 | −3.25002 |
| LCN2 |
7.38×10−254 |
2.67×10−257 | −3.10027 |
| TGM2 |
8.36×10−244 |
3.63×10−247 | −2.45302 |
| SAT1 |
3.53×10−238 |
1.79×10−241 | −2.46616 |
|
PDZK1IP1 |
3.63×10−230 |
2.11×10−233 | −4.94302 |
| PLAU |
7.60×10−226 |
4.96×10−229 | −2.07647 |
| SAA2 |
6.42×10−194 |
4.65×10−197 | −7.47593 |
| TNIP1 |
2.23×10−189 |
1.78×10−192 | −2.05325 |
| SPP1 |
2.23×10−189 |
1.94×10−192 | 2.29076 |
| S100A8 |
1.27×10−168 |
1.20×10−171 | −3.79768 |
|
TMEM132A |
1.42×10−166 |
1.44×10−169 | −1.93297 |
| ASNS |
6.48×10−156 |
7.04×10−159 | −2.09409 |
|
SERPINE1 |
2.92×10−149 |
3.38×10−152 | −2.55607 |
| PHLDB2 |
4.19×10−147 |
5.17×10−150 | −1.54635 |
| ICAM1 |
7.56×10−144 |
9.87×10−147 | −2.03788 |
| CXCL8 |
1.08×10−138 |
1.48×10−141 | −5.25318 |
Enriched GO terms and KEGG pathways of
DEGs and ASGs
The DEGs were enriched in 338 GO terms and 21 KEGG
pathways. The ASGs were enriched in 470 GO terms and 22 KEGG
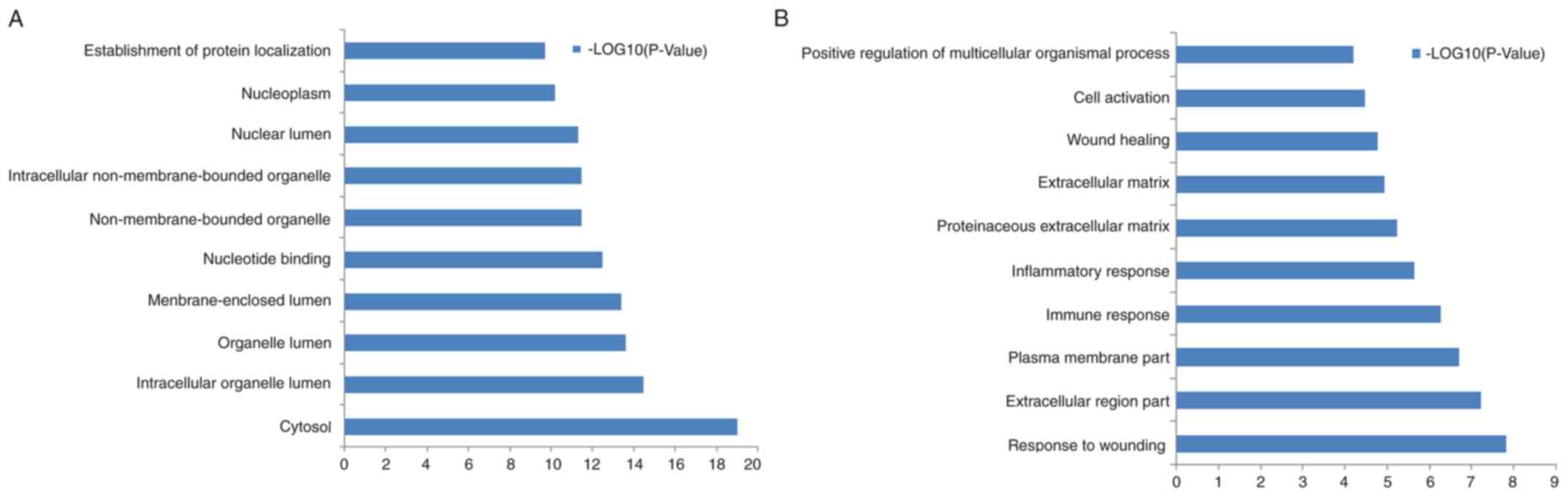
pathways. The top 10 GO terms for the ASGs and DEGs are listed in
Fig. 2A and B, respectively. Table III lists the enriched KEGG pathways
for the ASGs and DEGs. The DEGs were predominately enriched in
‘graft-vs.-host disease’, ‘hematopoietic cell lineage’,
‘ECM-receptor interaction’ and ‘NOD-like receptor signaling
pathway’. The ASGs were predominately enriched in ‘ubiquitin
mediated proteolysis’, ‘chronic myeloid leukemia’, ‘aminoacyl-tRNA
biosynthesis’ and ‘mTOR signaling pathway’.
 | Table III.Continued. |
Table III.
Continued.
| A, Enriched KEGG
pathways for DEGs |
|---|
|
|---|
| Pathway name | Genes, n | P-value |
|---|
| Graft-vs.-host
disease | 12 | 0.0006 |
| Hematopoietic cell
lineage | 18 | 0.0019 |
| NOD-like receptor
signaling pathway | 14 | 0.0038 |
| ECM-receptor
interaction | 17 | 0.0038 |
| Cell adhesion
molecules | 23 | 0.0044 |
| Allograft
rejection | 10 | 0.0045 |
| Cytokine-cytokine
receptor interaction | 38 | 0.0055 |
| Glycine, serine and
threonine metabolism | 9 | 0.0059 |
| MAPK signaling
pathway | 38 | 0.0075 |
| p53 signaling
pathway | 14 | 0.0085 |
| Natural killer cell
mediated cytotoxicity | 22 | 0.0100 |
| Toll-like receptor
signaling pathway | 18 | 0.0106 |
| Viral
myocarditis | 14 | 0.0122 |
| Nitrogen
metabolism | 7 | 0.0157 |
| Arginine and
proline metabolism | 11 | 0.0217 |
| Complement and
coagulation cascades | 13 | 0.0230 |
| Pathways in
cancer | 42 | 0.0266 |
| Axon guidance | 20 | 0.0274 |
| Pathogenic
Escherichia coli infection | 11 | 0.0344 |
| B cell receptor
signaling pathway | 13 | 0.0413 |
| Small cell lung
cancer | 14 | 0.0438 |
|
| B, Enriched KEGG
pathways for ASGs |
|
| Pathway
name | Gene, n | P-value |
|
| Ubiquitin mediated
proteolysis | 33 | 0.0002 |
| Chronic myeloid
leukemia | 20 | 0.0014 |
| Aminoacyl-tRNA
biosynthesis | 13 | 0.0029 |
| mTOR signaling
pathway | 15 | 0.0032 |
| Renal cell
carcinoma | 18 | 0.0040 |
| Pancreatic
cancer | 18 | 0.0054 |
| Neurotrophin
signaling pathway | 26 | 0.0079 |
| Pathways in
cancer | 55 | 0.0133 |
| Ribosome | 19 | 0.0173 |
| Pyrimidine
metabolism | 20 | 0.0208 |
| Acute myeloid
leukemia | 14 | 0.0215 |
| Small cell lung
cancer | 18 | 0.0250 |
| Wnt signaling
pathway | 28 | 0.0276 |
| Cell cycle | 24 | 0.0297 |
| VEGF signaling
pathway | 16 | 0.0373 |
| Lysine
degradation | 11 | 0.0377 |
| Insulin signaling
pathway | 25 | 0.0387 |
| Glioma | 14 | 0.0403 |
| Prostate
cancer | 18 | 0.0414 |
| Lysosome | 22 | 0.0465 |
| Endometrial
cancer | 12 | 0.0484 |
| N-glycan
biosynthesis | 11 | 0.0495 |
PPI network
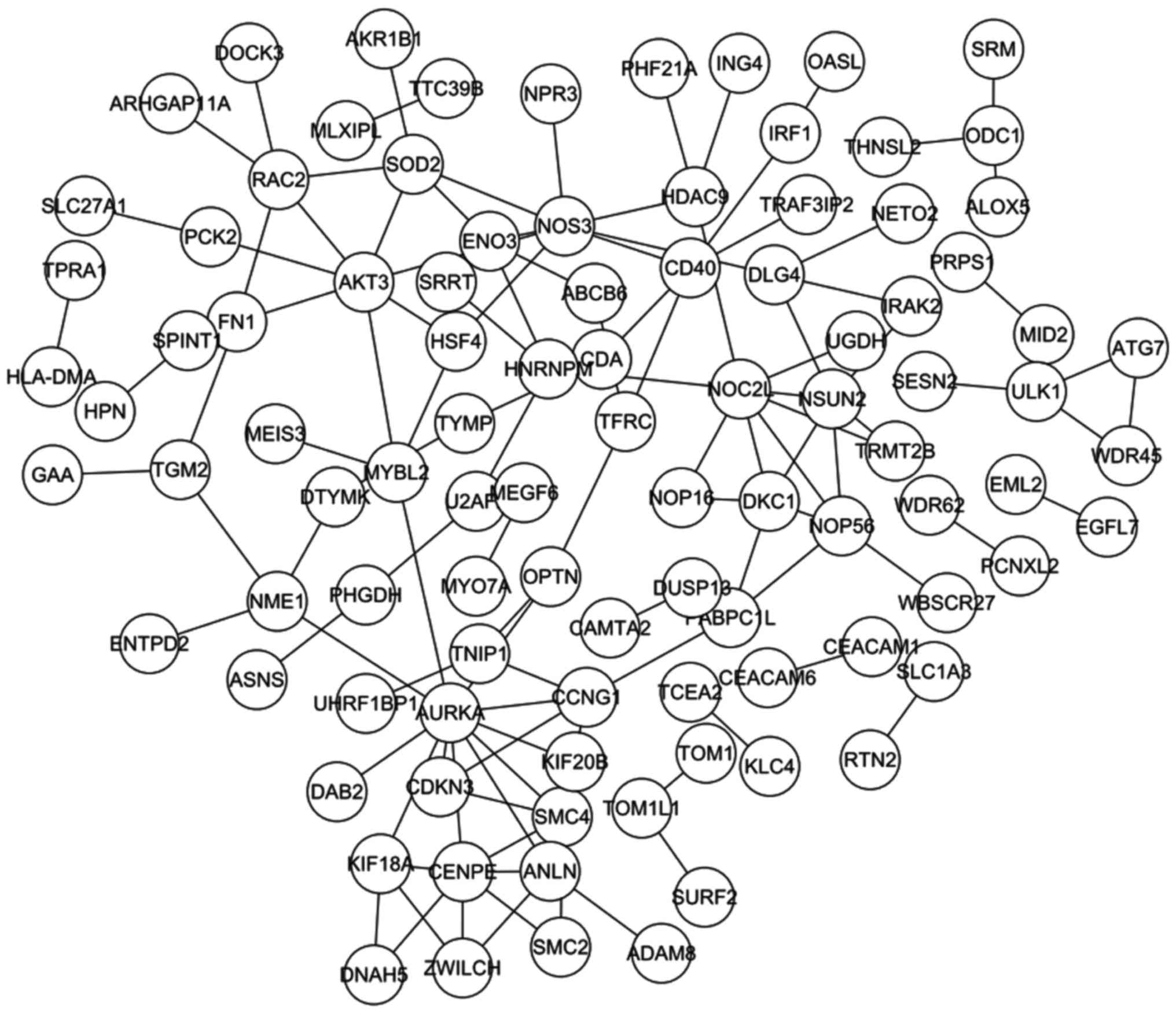
A total of 199 overlaps between the DEGs and the
ASGs were identified, and the PPI network constructed from the 199
overlapping genes contained 97 nodes and 115 pairs (Fig. 3). Table
IV lists the top 20 pairs with highest combined scores, and
Table V lists the top 10 nodes
according to connectivity degree.
 | Table IV.Top 20 pairs of the protein-protein
interaction network as determined by the highest combined
score. |
Table IV.
Top 20 pairs of the protein-protein
interaction network as determined by the highest combined
score.
| Gene 1 | Gene 2 | Combined score |
|---|
| SMC4 | SMC2 | 0.999 |
| NOP56 | DKC1 | 0.997 |
| TFRC | OPTN | 0.994 |
| SRM | ODC1 | 0.989 |
| CDA | TYMP | 0.987 |
| HNRNPM | U2AF2 | 0.979 |
| ZWILCH | CENPE | 0.977 |
| CEACAM6 | CEACAM1 | 0.970 |
| TGM2 | FN1 | 0.970 |
| NOS3 | AKT3 | 0.964 |
| DTYMK | NME1 | 0.954 |
| CENPE | KIF18A | 0.946 |
| NOP56 | NOC2L | 0.941 |
| ATG7 | ULK1 | 0.937 |
|
ARHGAP11A | RAC2 | 0.936 |
| ZWILCH | KIF18A | 0.930 |
| OASL | IRF1 | 0.925 |
| HDAC9 | PHF21A | 0.925 |
| HDAC9 | NOS3 | 0.923 |
| DKC1 | NOC2L | 0.911 |
 | Table V.The top 10 nodes of the
protein-protein interaction network as determined by the highest
connectivity degree. |
Table V.
The top 10 nodes of the
protein-protein interaction network as determined by the highest
connectivity degree.
| Gene | Degree |
|---|
| AURKA | 11 |
| NOS3 | 8 |
| NOC2L | 8 |
| CENPE | 7 |
| AKT3 | 7 |
| NSUN2 | 6 |
| SOD2 | 5 |
| SMC4 | 5 |
| RAC2 | 5 |
| NOP56 | 5 |
Discussion
Lung cancer is a serious threat to human health and
survival (22). Despite progress in
diagnosis and treatment, the 5-year survival rate of patients with
lung cancer is only 9–20% (23).
FOXD3 has been suggested to be a tumor suppressor in various
types of cancer (8–10). However, the underlying mechanism of
FOXD3 activity in lung cancer remains unclear. In the
present study, DEGs and ASGs between FOXD3-knockout and
normal lung cancer A549 cells were identified, and functional
enrichment analysis was performed to identify the associated
biological processes involved in lung cancer. Finally, a PPI
network of the most significant genes was constructed. These
results may contribute to the understanding of the role of
FOXD3 in lung cancer.
The most enriched GO terms for the DEGs were
‘response to wounding’, ‘extracellular region’, ‘plasma membrane’
and ‘immune response’. The ASGs were mainly enriched in ‘cytosol’,
‘intracellular organelle lumen’, ‘organelle lumen’ and
‘membrane-enclosed lumen’ (Fig. 2).
The wound response involves clotting and coagulation, tissue
remodeling, cellular migration and proliferation, and angiogenesis
(24). The majority of these
processes also serve important roles in the progression of cancer.
One study reported that the upregulation of factors associated with
the ‘wound response’ term was highly prognostic of breast cancer
survival, and revealed a strong association between the pathogenic
conditions identified by this signature and those identified using
serum-treated fibroblasts (25). In
lung cancer, the upregulation of genes associated with the ‘wound
response’ term has been demonstrated as predictive of poor overall
survival time and increased risk of metastasis (26).
The cell membrane is a biological membrane that
separates the interior of cells from the outside environment
(27). Plasma membrane fluidity
depends on the composition of the lipids and proteins in the
membrane, and has been demonstrated to be significantly associated
with the malignant potential of cancer cells (28), with alterations in the plasma membrane
fluidity of cancer cells associated with their capacity to form
metastases (29). In lung cancer,
studies reported that patients with high plasma membrane fluidity
had poorer prognoses than those with less fluid membranes, and the
fluidity variable may be used as an independent additional
prognostic factor (28,30,31).
Cytosol is the fluid within cells, a component
essential to the process of cytokinesis, a critical stage in cell
proliferation (32,33). Another major function of cytosol is to
transport metabolites; most tumor cells demonstrate different
metabolic pathways to normal cells (34). One study indicated that metabolism
contributed to the tumor proliferation, migration, and metastasis
of lung cancer (35).
Other enriched GO terms, e.g., ‘organelle lumen’,
have also been associated with tumorigenesis. Jingye et al
(36) reported that a disordered pH
in the organelle lumen is a common characteristic of cancer cells.
Despite a number of studies reporting the FOXD3-mediated
inhibition of the growth, invasion and migration of tumor cells in
various types of cancer, including lung cancer (37–39),
limited data is available regarding the association between
FOXD3 and these GO terms. As discussed, the identified GO
terms have been associated with the growth, invasion and migration
of tumor cells, thus it is speculated that FOXD3 may affect
the progression of lung cancer indirectly by regulating these
biological processes.
From the identified KEGG pathways, the mechanistic
target of rapamycin (mTOR) signaling pathway has also been
associated with the growth and proliferation of tumor cells, and
the deregulation of multiple elements of the mTOR pathway has been
reported in numerous types of cancer (40). The NOD-like receptor signaling pathway
is involved in the formation of inflammasomes, and numerous types
of cancer are associated with inflamed tissue (41). However, the associations between
FOXD3 and the identified KEGG pathways require further
exploration.
A total of 199 overlaps between the DEGs and the
ASGs were identified, from which the PPI network was constructed
(Fig. 3). The top 5 nodes of the PPI
network, with the highest degree, were aurora kinase A
(AURKA), nitric oxide synthase 3 (NOS3), NOC2-like
nucleolar associated transcriptional repressor (NOC2L),
centromere protein E (CENPE) and AKT3. The majority
of these genes have been previously associated with tumorigenesis.
AURKA and NOS3 serve important roles in the
development of various types of cancer, including lung cancer;
AURKA is a cell cycle-regulated kinase involved in spindle
formation and chromosome segregation (42). Various types of cancer exhibit the
overexpression of AURKA, which is associated with
chromosomal instability, centrosomal amplification/aneuploidy,
therapeutic resistance, cell-cycle progression and anti-apoptosis.
As an oncogene, AURKA is an important therapeutic target in
lung cancer, and cell proliferation, apoptosis and cell cycle
progression are associated with the expression of AURKA
(43). NOS3 encodes an enzyme
that regulates the production of nitric oxide and contributes to
uncontrollable cell growth in a number of cancer types (44). Various studies have demonstrated
associations between NOS3 and cancer processes. For example,
Arıkan et al (45) reported
that the NOS3 Glu298Asp polymorphism may be associated with
the risk and progression of colorectal cancer. Lee et al
(46) reported that genetic
polymorphisms in NOS3 modified individual susceptibility to
invasive breast cancer with lymph node involvement in Korean women.
Furthermore, the expression of NOS3 has been reported to
contribute to the tumor angiogenesis and lymph metastasis of human
non-small cell lung cancer (47).
The expression of other genes, including CENPE,
NOC2L and AKT3 has also been associated with
tumorigenesis (48–50). CENPE was identified as a novel
therapeutic candidate in neuroblastoma (50), and the selective activation of the
AKT3 protein promoted cell survival and tumor development in
non-familial melanomas in one study (48). To the best of our knowledge, there is
no experimental evidence of the direct association between
FOXD3 and these genes. However, the biological functions
associated with these genes in the context of cancer correspond
with the regulating mechanism of FOXD3 in lung cancer.
FOXD3 acts as a tumor suppressor by regulating the
expression of the target genes, thus inhibiting the growth,
invasion and migration of tumor cells (51). Few specific targets for FOXD3
in lung cancer have been reported, whereas AURKA and
NOS3 serve critical roles in the growth, invasion and
migration of tumor cells in lung cancer. Therefore, we speculate
that AURKA and NOS3 may be the targets of
FOXD3 that execute its effect in lung cancer. Confirmation
of these conclusions and further exploration of the specific
mechanism of FOXD3 regulation in lung cancer are
required.
In conclusion, FOXD3 serves an important role
in regulating the growth, migration and proliferation of lung
cancer cells. Genes such as AURKA and NOS3 may be
targets of FOXD3, mediating its effect in lung cancer. The
present study contributes to the existing understanding of the
molecular mechanism of lung cancer and may provide data to
contribute towards novel strategies for improving the diagnosis and
therapy of lung cancer.
Acknowledgements
The present study was supported by the Municipal
Science and Technology Commission of Tianjin (grant nos.
15ZLZLZF00440 and 16ZLZXZF00120) and the Health Bureau Science and
Technology Foundation of Tianjin (grant no. 2014KZ102).
References
|
1
|
Reck M, Heigener DF, Mok T, Soria JC and
Rabe KF: Management of non-small-cell lung cancer: Recent
developments. Lancet. 382:709–719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Grunnet M and Sorensen JB:
Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung
Cancer. 76:138–143. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lozano R, Naghavi M, Foreman K, Lim S,
Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et
al: Global and regional mortality from 235 causes of death for 20
age groups in 1990 and 2010: A systematic analysis for the Global
Burden of disease study 2010. Lancet. 380:2095–2128. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Y, Chen J, Wu S, Hu C, Li X, Wang Y,
Yang Y, Rajan N, Chen Y, Chen Y, et al: Clinical effectiveness and
clinical toxicity associated with platinum-based doublets in the
first-line setting for advanced non-squamous non-small cell lung
cancer in Chinese patients: A retrospective cohort study. BMC
Cancer. 14:9402014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cooper WA, Lam DC, O'Toole SA and Minna
JD: Molecular biology of lung cancer. Lung Cancer. 42:378–386.
2004.
|
|
6
|
Weigel D and Jäckle H: The fork head
domain: A novel DNA binding motif of eukaryotic transcription
factors? Cell. 63:455–456. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sutton J, Costa R, Klug M, Field L, Xu D,
Largaespada DA, Fletcher CF, Jenkins NA, Copeland NG, Klemsz M and
Hromas R: Genesis, a winged helix transcriptional repressor with
expression restricted to embryonic stem cells. J Biol Chem.
271:23126–23133. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yan JH, Zhao CL, Ding LB and Zhou X: FOXD3
suppresses tumor growth and angiogenesis in non-small cell lung
cancer. Biochem Biophys Res Commun. 466:111–116. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cheng AS, Li MS, Kang W, Cheng VY, Chou
JL, Lau SS, Go MY, Lee CC, Ling TK, Ng EK, et al: Helicobacter
pylori causes epigenetic dysregulation of FOXD3 to promote gastric
carcinogenesis. Gastroenterology. 144:122–133.e9. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Abel EV and Aplin AE: FOXD3 is a mutant
B-RAF-regulated inhibitor of G(1)-S progression in melanoma cells.
Cancer Res. 70:2891–2900. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Katiyar P and Aplin AE: FOXD3 regulates
migration properties and Rnd3 expression in melanoma cells. Mol
Cancer Res. 9:545–552. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li D, Mei H, Qi M, Yang D, Zhao X, Xiang
X, Pu J, Huang K, Zheng L and Tong Q: FOXD3 is a novel tumor
suppressor that affects growth, invasion, metastasis and
angiogenesis of neuroblastoma. Oncotarget. 4:2021–2044. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim D, Pertea G, Trapnell C, Pimentel H,
Kelley R and Salzberg SL: TopHat2: Accurate alignment of
transcriptomes in the presence of insertions, deletions and gene
fusions. Genome Biol. 14:R362013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Anders S, Pyl PT and Huber W: HTSeq-a
Python framework to work with high-throughput sequencing data.
Bioinformatics. 31:166–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Culpepper SA and Aguinis HR: Is for
Revolution: A Cutting-Edge, free, open source statistical package.
Organizational Res Methods. 13:735–740. 2011. View Article : Google Scholar
|
|
16
|
Warnes GR, Bolker B, Bonebakker L, et al:
Gplots: Various R programming tools for plotting data. R package
version 2.12. 1. http://CRAN.R-project.org/package=gplots
|
|
17
|
Shen S, Park JW, Lu ZX, Lin L, Henry MD,
Wu YN, Zhou Q and Xing Y: rMATS: Robust and flexible detection of
differential alternative splicing from replicate RNA-Seq data. Proc
Natl Acad Sci USA. 111:pp. E5593–E5601. 2014; View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for Annotation,
Visualization, and Integrated Discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39(Database issue): D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41(Database issue): D808–D815. 2013.PubMed/NCBI
|
|
21
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang P: Epidemiology of lung cancer
prognosis: Quantity and quality of life. Methods Mol Biol.
471:469–86. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schäfer M and Werner S: Cancer as an
overhealing wound: An old hypothesis revisited. Nat Rev Mol Cell
Biol. 9:628–638. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Troester MA, Lee MM, Carter M, Fan C,
Cowan DW, Perez ER, Pirone JR, Perou CM, Jerry DJ and Schneider SS:
Activation of host wound responses in breast cancer
microenvironment. Clin Cancer Res. 15:7020–7028. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chang HY, Nuyten DS, Sneddon JB, Hastie T,
Tibshirani R, Sørlie T, Dai H, He YD, van't Veer LJ, Bartelink H,
et al: Robustness, scalability, and integration of a wound-response
gene expression signature in predicting breast cancer survival.
Proc Natl Acad Sci USA. 102:pp. 3738–3743. 2005; View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Singleton P: Bacteria in Biology,
Biotechnology, and Medicine. 5th. John Wiley; Hoboken, NJ: pp.
444–454. 1999
|
|
28
|
Sok M, Sentjurc M, Schara M, Stare J and
Rott T: Cell membrane fluidity and prognosis of lung cancer. Ann
Thorac Surg. 73:1567–1571. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nakazawa I and Iwaizumi M: A role of the
cancer cell membrane fluidity in the cancer metastases: An ESR
study. Tohoku J Exp Med. 157:193–198. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Deliconstantinos G: Physiological aspects
of membrane lipid fluidity in malignancy. Anticancer Res.
7:1011–1021. 1987.PubMed/NCBI
|
|
31
|
Nakazawa I and Iwaizumi M: A role of the
cancer cell membrane fluidity in the cancer metastases: Aan ESR
study. Tohoku J Exp Med. 157:193–198. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Winey M, Mamay CL, O'Toole ET, Mastronarde
DN, Giddings TH Jr, McDonald KL and McIntosh JR: Three-dimensional
ultrastructural analysis of the Saccharomyces cerevisiae mitotic
spindle. J Cell Biol. 129:1601–1615. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jana SS, Kawamoto S and Adelstein RS: A
specific isoform of nonmuscle myosin II-C is required for
cytokinesis in a tumor cell line. J Biol Chem. 281:24662–24670.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Weisiger RA: Cytosolic fatty acid binding
proteins catalyze two distinct steps in intracellular transport of
their ligands. Mol Cell Biochem. 239:35–43. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li XB, Gu JD and Zhou QH: Review of
aerobic glycolysis and its key enzymes-new targets for lung cancer
therapy. Thoracic Cancer. 6:17–24. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jingye Z, Zining L, Peng L, Jun Q, Xinwei
L, Lu W, Wei F, Liang C, Xunbin W and Cong L: Selective imaging and
cancer cell death via pH switchable near-infrared fluorescence and
photothermal effects. Chem Sci. 7:5995–6005. 2016. View Article : Google Scholar
|
|
37
|
Ackermann S, Kocak H, Hero B, Ehemann V,
Kahlert Y, Oberthuer A, Roels F, Theißen J, Odenthal M, Berthold F
and Fischer M: FOXP1 inhibits cell growth and attenuates
tumorigenicity of neuroblastoma. Bmc Cancer. 14:8402014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kos R, Reedy MV, Johnson RL and Erickson
CA: The winged-helix transcription factor FoxD3 is important for
establishing the neural crest lineage and repressing melanogenesis
in avian embryos. Development. 128:1467–1479. 2001.PubMed/NCBI
|
|
39
|
Wang C, Huang Y and Dai W: Tumor
suppression function of FoxD3 in lung cancer. Ir J Med Sci.
185:547–553. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pópulo H, Lopes JM and Soares P: The mTOR
signalling pathway in human cancer. Int J Mol Sci. 13:1886–1918.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Castaño-Rodríguez N, Kaakoush NO, Goh KL,
Fock KM and Mitchell HM: The NOD-like receptor signalling pathway
in Helicobacter pylori infection and related gastric cancer: A
case-control study and gene expression analyses. PLoS One.
9:e988992014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lens SM, Voest EE and Medema RH: Shared
and separate functions of polo-like kinases and aurora kinases in
cancer. Nat Rev Cancer. 10:825–841. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ma ZL, Zhang BJ, Wang DT, Li X, Wei JL,
Zhao BT, Jin Y, Li YL and Jin YX: Tanshinones suppress AURKA
through up-regulation of miR-32 expression in non-small cell lung
cancer. Oncotarget. 6:20111–20120. 2015.PubMed/NCBI
|
|
44
|
Xu W, Charles IG, Moncada S, Gorman P,
Sheer D, Liu L and Emson P: Mapping of the genes encoding human
inducible and endothelial nitric oxide synthase (NOS2 and NOS3) to
the pericentric region of chromosome 17 and to chromosome 7,
respectively. Genomics. 21:419–422. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Arıkan S, Cacina C, Guler E, Çulcu S, Tuna
G and Yaylımeraltan I: The effects of NOS3 Glu298Asp variant on
colorectal cancer risk and progression in Turkish population. Mol
Biol Rep. 39:3245–3249. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lee KM, Choi JY, Lee JE, Noh DY, Ahn SH,
Han W, Yoo KY, Hayes RB and Kang D: Genetic polymorphisms of NOS3
are associated with the risk of invasive breast cancer with lymph
node involvement. Breast Cancer Res Treat. 106:433–438. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang JH, Chen LB, Heng-Hui MA and Meng K:
Expressions of NOS2 and NOS3 in human non-small cell lung cancer
and the relationship with tumor angiogenesis and lymph node
metastasis. J Med Postgraduates. 2004.
|
|
48
|
Stahl JM, Sharma A, Cheung M, Zimmerman M,
Cheng JQ, Bosenberg MW, Kester M, Sandirasegarane L and Robertson
GP: Deregulated Akt3 activity promotes development of malignant
melanoma. Cancer Res. 64:7002–7010. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Terasaka Y, Miyazaki D, Yakura K, Haruki T
and Inoue Y: Induction of IL-6 in transcriptional networks in
corneal epithelial cells after herpes simplex virus type 1
infection. Invest Ophthalmol Vis Sci. 51:2441–2449. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Balamuth NJ, Wood A, Wang Q, Jagannathan
J, Mayes P, Zhang Z, Chen Z, Rappaport E, Courtright J, Pawel B, et
al: Serial transcriptome analysis and cross-species integration
identifies centromere-associated protein E as a novel neuroblastoma
target. Cancer Res. 70:2749–2758. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Weiss MB, Abel EV, Dadpey N and Aplin AE:
FOXD3 modulates migration through direct transcriptional repression
of TWIST1 in melanoma. Mol Cancer Res. 12:1314–1323. 2014.
View Article : Google Scholar : PubMed/NCBI
|