Introduction
Breast cancer is the most common malignant tumor
among women and is a leading cause of cancer-associated death in
women worldwide. Based on gene expression patterns, breast cancer
was divided into five distinct molecular subtypes including luminal
A, luminal B, receptor tyrosine-protein kinase erbB-2
(HER2)-enriched, basal-like and normal-like subtype (1). Triple negative breast cancer (TNBC) is
defined as the absence of estrogen receptor (ER), progesterone
receptor (PR) and HER2 expression, accounting for approximately
15–20% of all breast cancer patients (2). The majority of TNBC patients (up to 70%)
overlap with the basal-like gene expression subtype (3). Compared with other breast cancer
subtypes, TNBC lacks clinically efficient targeted therapies and is
usually more aggressive with higher rates of distant metastasis and
poorer overall survival (2).
Therefore, it is imperative to better elucidate the underlying
mechanism of TNBC and identify novel targets for effective
therapies.
microRNAs (miRNAs) are a class of small non-coding
RNAs that are 19–25 nucleotides in size, which regulate gene
expression primarily through mRNA degradation or translational
repression. It has been estimated that approximately over one third
of human protein-coding genes appeared to be conserved miRNA
targets, which suggests that miRNAs may serve an important role in
gene expression (4). miRNAs are found
to be critically involved in various cancer-associated cell
processes, including proliferation, differentiation, cell-cycle
control, apoptosis, invasion and metastasis (5,6). miR-9 was
demonstrated to be associated with epithelial-mesenchymal
transition, aggressive phenotype and poor prognosis in breast
cancer, suggesting that miR-9 may serve as a promising biomarker
for breast cancer progression (7). It
was reported that miR-145 expression was downregulated in
MDA-MB-231 cells and overexpression of miR-145 inhibited tumor
invasion by targeting ARF6, a known regulator of breast tumor cell
invasion (8). However, the miRNA
expression profile in TNBC and the role of miRNAs in modulating the
pathways of TNBC remain largely unexplored.
In the present study, 107 aberrant miRNAs were
identified in human MDA-MB-231 breast cancer cells, compared with
MCF-7 cells by means of miRNA microarray analysis. Five prominently
dysregulated miRNAs (miR-200c-3p, miR-221-3p, miR-222-3p,
miR-192-5p and miR-146a) were further validated by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR). In
addition, bioinformatic analysis including gene ontology analysis
and pathway enrichment analysis, were performed in an attempt to
reveal the potential roles of these dysregulated miRNAs and
associated signaling pathways involved in development and
progression of TNBC.
Materials and methods
Cell culture
Human breast cancer cell lines MDA-MB-231 (ER, PR
and HER2 negative) and MCF-7 (ER, PR positive and HER2 negative)
were obtained from the Type Culture Collection of Chinese Academy
of Sciences (Shanghai, China). MDA-MB-231 cells were maintained in
Leibovitz's L-15 medium (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum
(Hyclone; GE Healthcare, Chicago, IL, USA), 100 U/ml penicillin and
100 µg/ml streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.). MCF-7 cells were cultured in RPMI-1640 medium (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(Hyclone; GE Healthcare), 100 U/ml penicillin and 100 µg/ml
streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.). Cell
lines were cultured and maintained at 37°C in a humidified
atmosphere containing 5% carbon dioxide.
miRNA microarray profiling
miRNA microarray analysis including separation,
quality control, labeling, hybridization and scanning was performed
by LC Sciences (Houston, TX, USA). The array contained 1090 human
mature miRNA probes for all human mature miRNAs based on Sanger
miRase Release 15.0 (The Wellcome Trust Sanger Institute, Hinton,
UK). Hybridization was performed on a µParaflo microfluidic chip
using a micro-circulation pump (Atactic Technologies, Inc.,
Houston, TX, USA). Images were collected using a laser scanner
(GenePix 4000B; Molecular Devices, LLC, Sunnyvale, CA, USA) and
digitized using Array-Pro image analysis software (Media
Cybernetics, Inc., Rockville, MD, USA). The data were analyzed by
first subtracting the background and then normalizing the signals
using a LOWESS filter (Locally-weighted Regression). Student's
t-test analysis was conducted to identify differentially expressed
miRNAs between MDA-MB-231 and MCF-7 cells. The false discovery rate
(FDR) was P<0.05 and P<0.01 and served as the cut-off
criteria. The data were log2 transformed and median centered by
Cluster 3.0 software (Informer Technologies Inc., Los Angeles, CA,
USA) and then further analyzed with hierarchical clustering with
average linkage.
RT-qPCR for miRNA expression
Total RNA from cultured cells was extracted using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The concentration and
quality of RNA was determined using the Nanodrop 1000
spectrophotometer (Thermo Fisher Scientific, Inc.). RNA (1 µg) was
reverse transcribed using Ncode™ miRNA First-Strand cDNA Synthesis
kit (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The forward primers used for amplification
were as follows: miR-200c-3p: 5′-UAAUACUGCCGGGUAAUGAUGGA-3′;
miR-221-3p: 5′-GCTACATTGTCTGCTGGGTTTC-3′; miR-222-3p:
5′-GCTACATCTGGCTACTGGGT-3′; miR-192-5p:
5′-GCTGACCTATGAATTGACAGCC-3′; miR-146a: 5′-CAGTGCGTGTCGTGGAGT-3′,
and U6 snRNA: 5′-GCTTCGGCAGCACATATACTAAAAT-3′. The universal
reverse primer was 5′-GAGACTGCGGATGTATAGAACTTGA-3′. Quantitative
PCR was performed using SYBR®Green Realtime PCR Master
mix kits (Toyobo Life Science, Osaka, Japan) on a Bio-Rad CFX384
PCR instrument (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
according to the manufacturer's protocol. The PCR thermocycling
conditions were as follows: Initial denaturation at 95°C for 10 min
followed by 40 cycles of 95°C for 15 sec and 63°C for 25 sec. U6
snRNA was used as an internal control. Each sample was run in
triplicate. The level of miRNA expression was measured using the
2−ΔΔCq method (9). The
results are presented as fold-change of each miRNA in the
MDA-MB-231 cells relative to the MCF-7 cells.
Target prediction and bioinformatic
analysis
Targets of miRNAs were predicted by using online
miRNA target prediction algorithms: TargetScan, PicTar4, and
miRanda (www.targetscan.org, http://pictar.mdc-berlin.de and www.microrna.org, respectively), which are commonly
used to predict the targets of miRNAs. The intersection of miRNA
target genes was selected for further analysis. Gene ontology (GO)
analysis was performed at three levels: Molecular function,
biological process and cellular component. Pathway analysis was
performed using the Kyoto Encyclopedia of Genes and Genomes
database (KEGG) (www.genome.jp/kegg). Functional enrichment and pathway
enrichment analyses were performed by using the Database for
Annotation, Visualization and Integrated Discovery (DAVID software;
http://david-d.ncifcrf.gov) (10). P-value was adjusted by the method of
Benjamini-Hochberg to control the false discovery rate (11). Enriched GO terms and KEGG pathways
were identified as significant with P<0.05. Cytoscape 2.8.3
(www.cytoscape.org) software was applied to
construct the possible functional network of the selected miRNAs
and targets (12).
Statistical analysis
Statistical analysis was performed by using the SPSS
version 16.0 statistical package (SPSS, Inc., Chicago, IL, USA)
using the Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of dysregulated miRNAs
in MDA-MB-231 cells
To investigate the miRNA profiles in triple negative
breast cancer, human breast cancer cell lines MDA-MB-231 (ER, PR
and HER2 negative) and MCF-7 (ER, PR positive and HER2 negative)
were selected. The miRNA expression profiles of MDA-MB-231 and
MCF-7 cells were evaluated by using miRNA microarray analysis. The
cluster analysis revealed that MDA-MB-231 cells were characterized
by significant alterations in miRNA expression compared with MCF-7
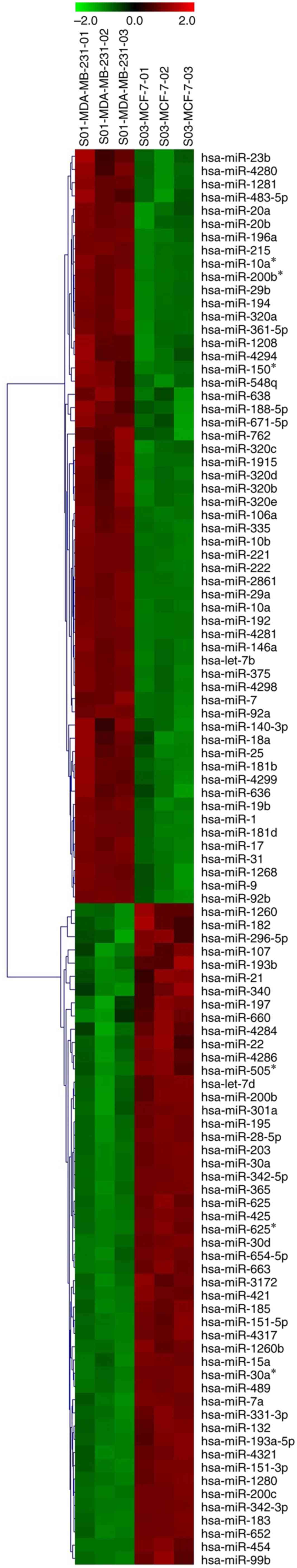
cells. As shown in Fig. 1, 107 miRNAs
were identified that were differentially expressed in MDA-MB-231
cells compared with MCF-7 cells (FDR<0.05; P<0.01). Among the
107 miRNAs, 57 miRNAs were upregulated, and 50 miRNAs were
downregulated in MDA-MB-231 cells.
Validation of miRNA expression by
RT-qPCR
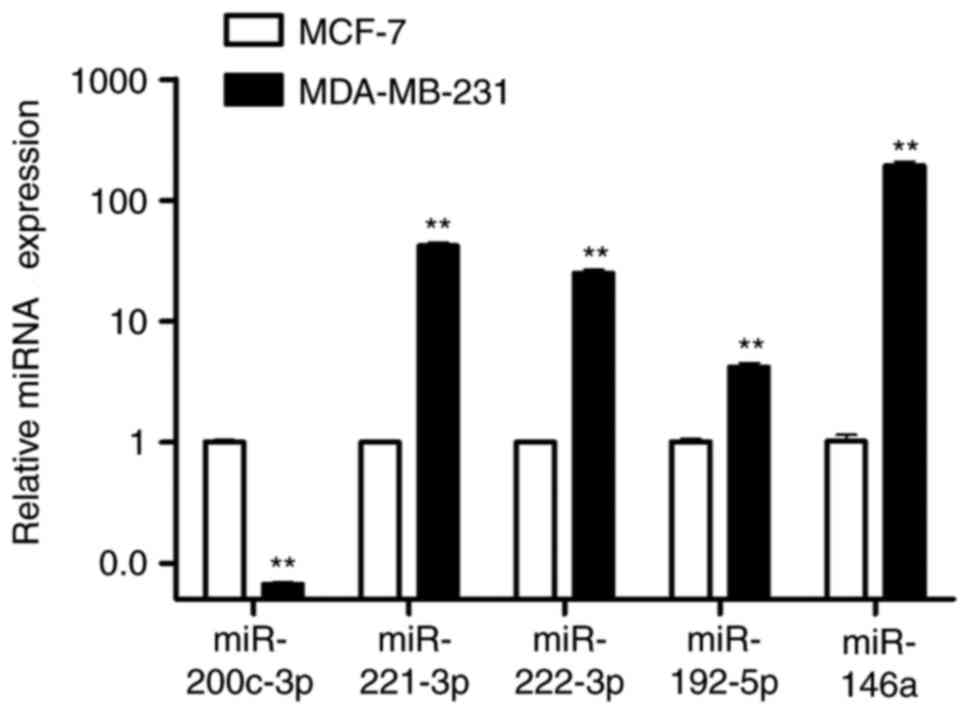
Of the 107 differentially expressed miRNAs obtained
from the microarray analysis, the top 5 dysregulated miRNAs
(miR-200c-3p, miR-221-3p, miR-222-3p, miR-192-5p and miR-146a) were
further verified by RT-qPCR. It was found that there was a
downregulation in miR-200c-3p expression and a significant
upregulation of miR-221-3p, miR-222-3p, miR-192-5p and miR-146a
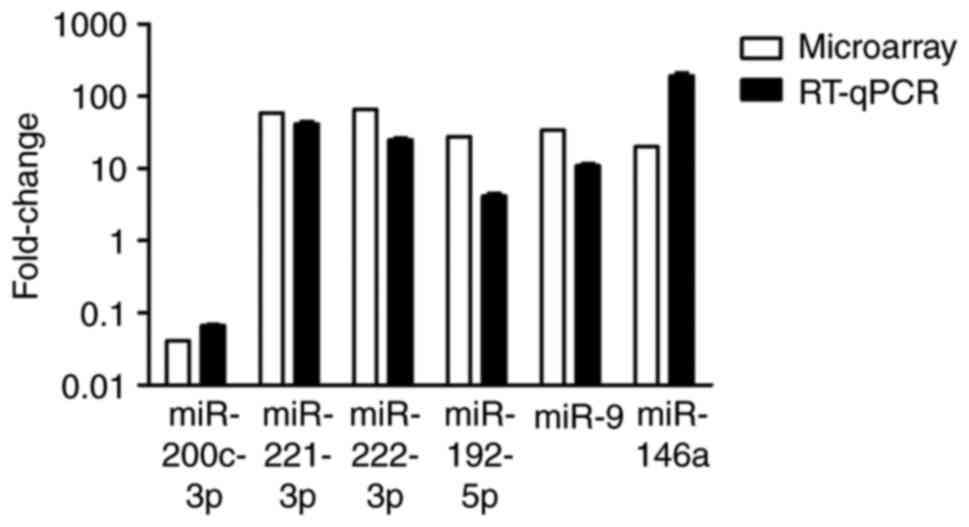
expression in MDA-MB-231 cells compared with MCF-7 cells (Fig. 2). Fig. 3
shows a comparison of microarray and RT-qPCR results for the five
miRNAs. The results of RT-qPCR consistently confirmed the results
obtained by microarray analysis.
GO analysis
To elucidate functions of dysregulated miRNAs in
TNBC, potential targets of five prominently dysregulated miRNAs
validated by RT-qPCR were predicted by miRNA target prediction
algorithms. A total of 597 target genes were identified for further
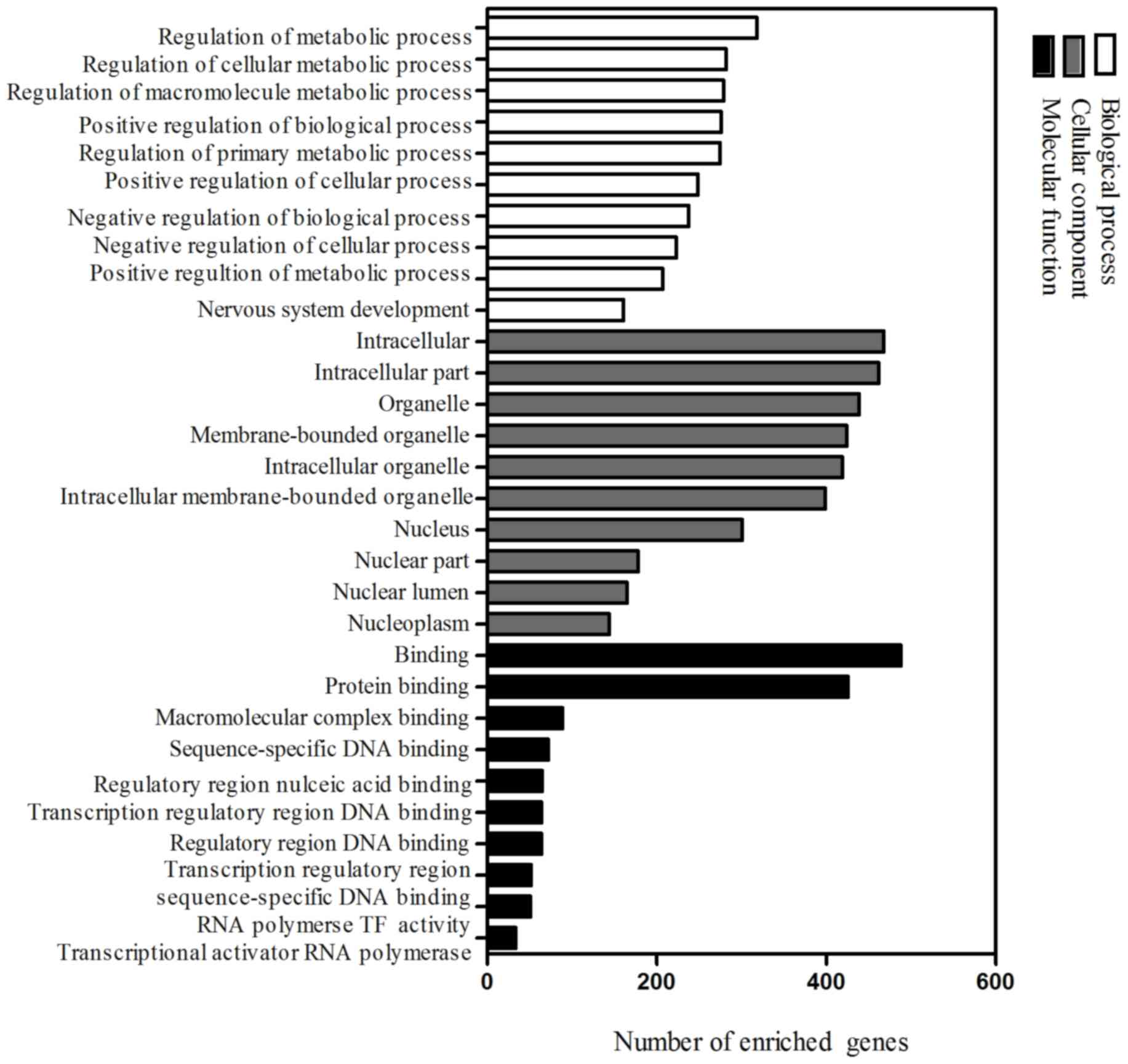
analysis. Functional enrichment analysis was performed using DAVID.
A total of 955, 84 and 60 GO items were obtained, respectively, in
the ontologies of biological process, cellular component and
molecular function (P<0.05). The 10 most significantly
associated terms for each ontology are demonstrated in Fig. 4. It was found that predicted targets
of selected miRNAs were involved in important processes such as
regulation of metabolic process, transcription factor activity, DNA
binding and protein binding.
KEGG pathway analysis and network
analysis
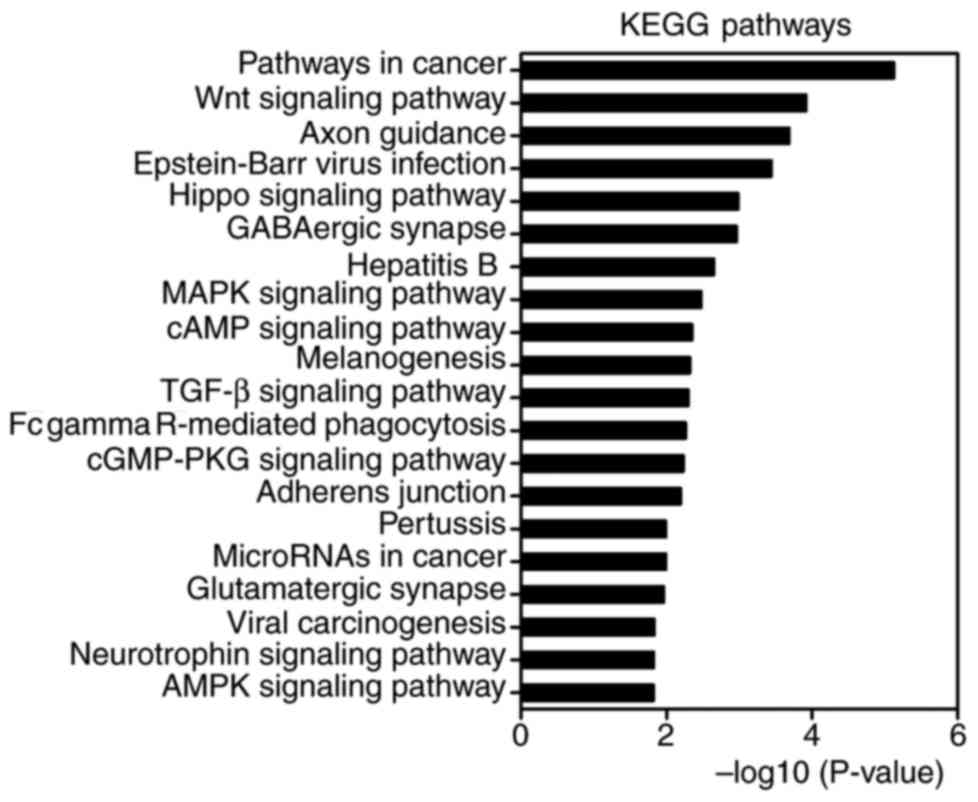
KEGG pathway enrichment analysis was performed by
using DAVID. There were 53 pathways obtained and 35 of them were
statistically significant (P<0.05). Of these, important pathways
involved in tumorigenesis and metastasis were enriched, including
the MAPK signaling pathway, Wnt signaling pathway, TGF-β signaling
pathway, pathways in cancer, cyclic adenosine monophosphate (cAMP)
signaling pathway, miRNAs in cancer, transcriptional dysregulation
in cancer, AMP-activated protein kinase (AMPK) signaling pathway,
ubiquitin mediated proteolysis, cell cycle, adherens junction and
gap junction. The top 20 highly enriched KEGG pathways are
presented in Fig. 5. To further
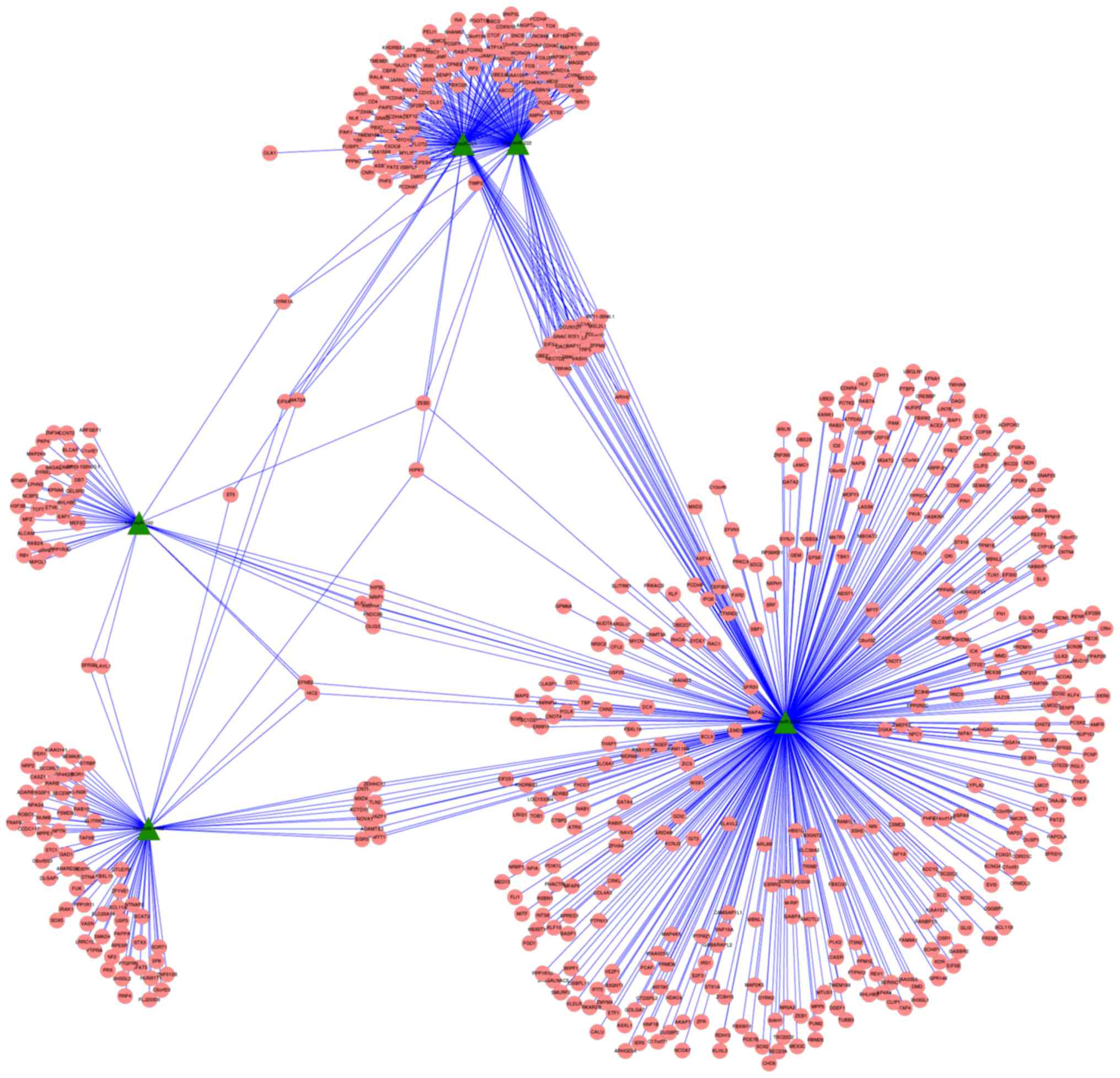
elucidate associations between selected dysregulated miRNAs and
potential target genes, miRNA-gene network analysis was constructed
by Cytoscape software (Fig. 6). As
shown in the network, miR-200c may regulate several hundreds of
target genes, suggesting that miR-200c may serve important roles in
breast cancer.
Discussion
Breast cancer is considered a complex and
heterogeneous disease with distinct molecular subtypes and
therapeutic responses. Although great improvements have been made
in the treatment of breast cancer, such as endocrine therapy and
HER2-targeted therapy, there are currently no available molecularly
targeted therapeutics for patients with TNBC. Emerging evidence has
demonstrated that miRNAs may serve as potential biomarkers and
promising therapeutic targets for breast cancer (13,14). In
this study, miRNA microarray analysis of MDA-MB-231 cells (TNBC
cells) and MCF-7 cells (breast adenocarcinoma luminal cells) was
performed. It was reported by Terao et al that differential
profiles of miRNA expression including miR-200c were determined in
MCF-7 and MDA-MB-231 cells (15). In
the present study, 107 miRNAs (57 upregulated and 50 downregulated)
were identified to be differentially expressed in MDA-MB-231 cells
compared with MCF-7 cells, suggesting that an altered expression of
miRNAs may contribute to TNBC progression.
In this study, five prominently dysregulated miRNAs
were further validated by RT-qPCR. The results were consistent with
the results obtained by microarray analysis. It was demonstrated
that miR-200c-3p expression levels in MDA-MB-231 cells were
downregulated compared with MCF-7 cells. miRNA-gene network
analysis also suggested that miR-200c shold be futher investigated
for it's potential role in breast cancer. miR-200c has been
demonstrated to serve important roles in the process of
epithelial-to-mesenchymal transition by directly targeting the
transcription factors zinc-finger E-box binding homeobox (ZEB)1 and
ZEB2 (16,17), two of the target genes in the analysis
of the present study. miR-200c expression may induce a significant
reorganization of tumor architecture, affecting important processes
involved in cell adhesion and motility. Induced miR-200c expression
significantly decreased the metastatic potential of a p53-knockout
and low-claudin expressing tumor model with highly aggressive
characteristics (18). miR-200c
expression levels from TNBC patients was significantly reduced in
cancer tissues compared with matched normal adjacent tissues by
qPCR analysis (19). Overexpression
of miR-200c decreased cell proliferation and promoted cell
apoptosis in MDA-MB-231 cells by targeting the expression of
X-linked inhibitor of apoptosis (19). It was reported that basal like breast
tumor subtypes had lower levels of the miR-200 family compared with
luminal subtypes (20), which was
similar to the findings of the present study. In a previous study,
it was found that the downregulation of miR-200c was associated
with poor chemotherapy response in human breast cancer patients and
the upregulation of miR-200c enhanced chemosensitivity to
epirubicin partially via regulation of multi-drug resistance gene
1/P-glycoprotein expression (21).
miR-200c was demonstrated to increase anoikis sensitivity by
targeting nuclear factor-κB and upregulated the tropomycin receptor
kinase B/neutrophin 3 autocrine signaling loop in triple negative
breast cancer (22). The miR-200 and
miR-221 families were revealed to exert opposing effects on
cellular plasticity during breast tumorigenesis (23). High miR-200 family expression promoted
a well-differentiated epithelial phenotype whereas overexpression
of miR-221/222 resulted in a poorly differentiated mesenchymal-like
phenotype (23). A previous study
demonstrated that knockdown of miR-221 inhibited cell migration and
invasion by altering E-cadherin expression in TNBC cells (24). Søkilde et al (25) reported that there was an inverse
association between miR-221/miR-222 and estrogen receptor
expression by microarray analysis in luminal and basal breast
cancer cell lines. A similar result was also found in highly
invasive basal-like breast cancer cells and non-invasive luminal
cells (26). It was demonstrated that
miR-221/222 promoted S-phase entry and cellular migration in
basal-like breast cancer (26). In
this study, miR-221 and miR-222 expression levels were measured by
RT-qPCR and were elevated in triple-negative type cells, which
suggested that miR-221 and miR-222 may serve an important role in
tumorigenesis in TNBC. In addition, it was found that miR-146a
expression was significantly upregulated in MDA-MB-231 cells
compared with MCF-7 cells. It was demonstrated that miR-146a
downregulated BRCA1 expression, as confirmed by reporter assays
(27). Another study reported that
knockdown of miR-146a in BRCA1-overexpressing cells suppressed the
ability to inhibit proliferation and transformation (28).
By using GO analysis, it was revealed that the
target genes of aberrant miRNAs were associated with important
processes such as regulation of metabolic processes, transcription
factor activity and DNA binding. According to KEGG pathway
enrichment analysis by using DAVID, the present study demonstrated
that the most significant pathways included the MAPK signaling
pathway, Wnt signaling pathway, TGF-β signaling pathway, cAMP
signaling pathway, AMPK signaling pathway, ubiquitin mediated
proteolysis, cell cycle, adherens junction and gap junction, which
were closely associated with tumor progression and metastasis.
Increasing evidence has demonstrated that various and complex
molecular signaling pathways were involved in TNBC progression and
metastasis. The MAPK signaling pathway serves an important role in
the regulation of cell proliferation, and aberrant activity of MAPK
has been implicated in the development and progression of TNBCs
(29). In a study involving 75 cases
of TNBC patients with lymph-node metastases, it was demonstrated
that phosphorylated extracellular-signal regulated kinase (pERK)
expression was associated with longer event-free survival (duration
from surgery until recurrence or mortality) and overall survival
(duration from surgery until the date of mortality or the most
recent date of follow-up) in TNBC, and pERK expression was an
independent prognostic factor by multivariate analysis (30). Based upon gene expression profiles,
diverse TNBC subtypes including basal-like 1 and basal-like 2,
immunomodulatory, mesenchymal, mesenchymal stem-like and luminal
androgen receptor were identified (31). The mesenchymal and mesenchymal
stem-like subtypes were enriched in signaling pathways involving
TGF-β, mechanistic target of rapamycin, Wnt/β-catenin,
platelet-derived growth factor receptor, and vascular endothelial
growth factor (31). It was found
that treatment with Wnt pathway inhibitors in TNBC resulted in
increased apoptosis and decreased cell proliferation and migration,
which suggested that targeting the Wnt pathway may have therapeutic
benefit for TNBC patients (32). It
was reported that upregulation and enrichment of TGF-β target genes
were most frequent in mesenchymal stem-like TNBC or TNBC with low
expression of claudin (33). TGF-β
signaling resulted in increased cell proliferation, migration,
invasion and motility, whereas these effects were abrogated by a
specific inhibitor against TGF-β receptor I and the anti-diabetic
agent metformin in breast cancer cell lines (33). In addition, a high expression of TGF-β
target genes were demonstrated to be associated with poor prognosis
in claudin-low patients (33). The
findings of the present study suggested that several signaling
pathways are regulated by miRNAs including the MAPK pathway, Wnt
pathway and TGF-β pathway, and may serve significant roles in the
development and progression of TNBC. Targeting these signaling
pathways may have the potential to serve as a novel and promising
strategy for the treatment of patients with TNBC.
In conclusion, 107 aberrant miRNAs in TNBC cells
were identified using miRNA microarray analysis, and 597 potential
target genes of five prominently dysregulated miRNAs that were
validated by RT-qPCR were selected by miRNA target prediction
algorithms. In addition, bioinformatic analysis suggested that
certain pathways such as the MAPK, Wnt and TGF-β signaling pathways
were primarily involved in tumorigenesis and metastasis in TNBC.
miRNA-gene network analysis suggested that miR-200c may contribute
to breast cancer development. Investigating and validating key
miRNAs and associated signaling pathways in TNBC patients compared
with non-TNBC patients may be warranted in future work. Taken
together, these findings may provide a view of the function of
dysregulated miRNAs in TNBC, suggesting that dysregulated miRNAs
may serve as potential biomarkers and therapeutic targets in
TNBC.
Acknowledgements
The present study was supported by the grants from
the Natural Science Foundation of Zhejiang Province (grant no.
LQ13H160016), the Medical Science and Technology Program of
Zhejiang Province (grant nos. 2013KYA026 and 2015DTA004) to JQ
Chen, the Foundation of Science and Technology Department of
Zhejiang Province (grant no. 2013C33205), the Medical Science and
Technology Program of Zhejiang Province (grant no. 2013KYB034) to
ZH Chen and the National Nature Science Foundation of China (grant
no. 81672597) to XJ Wang.
References
|
1
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arnedos M, Bihan C, Delaloge S and Andre
F: Triple-negative breast cancer: Are we making headway at least.
Ther Adv Med Oncol. 4:195–210. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Izumiya M, Tsuchiya N, Okamoto K and
Nakagama H: Systematic exploration of cancer-associated microRNA
through functional screening assays. Cancer Sci. 102:1615–1621.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gwak JM, Kim HJ, Kim EJ, Chung YR, Yun S,
Seo AN, Lee HJ and Park SY: MicroRNA-9 is associated with
epithelial-mesenchymal transition, breast cancer stem cell
phenotype, and tumor progression in breast cancer. Breast Cancer
Res Treat. 147:39–49. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Eades G, Wolfson B, Zhang Y, Li Q, Yao Y
and Zhou Q.: lincRNA-RoR and miR-145 regulate invasion in
triple-negative breast cancer via targeting ARF6. Mol Cancer Res.
13:330–338. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yan LX, Huang XF, Shao Q, Huang MY, Deng
L, Wu QL, Zeng YX and Shao JY: MicroRNA miR-21 overexpression in
human breast cancer is associated with advanced clinical stage,
lymph node metastasis and patient poor prognosis. RNA.
14:2348–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang X, Peng Y, Jin Z, Huang W, Cheng Y,
Liu Y, Feng X, Yang M, Huang Y, Zhao Z, et al: Integrated miRNA
profiling and bioinformatics analyses reveal potential causative
miRNAs in gastric adenocarcinoma. Oncotarget. 6:32878–32889. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kleivi Sahlberg K, Bottai G, Naume B,
Burwinkel B, Calin GA, Børresen-Dale AL and Santarpia L: A serum
microRNA signature predicts tumor relapse and survival in
triple-negative breast cancer patients. Clin Cancer Res.
21:1207–1214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Humphries B, Wang Z, Oom AL, Fisher T, Tan
D, Cui Y, Jiang Y and Yang C: MicroRNA-200b targets protein kinase
Cα and suppresses triple-negative breast cancer metastasis.
Carcinogenesis. 35:2254–2263. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Terao M, Fratelli M, Kurosaki M, Zanetti
A, Guarnaccia V, Paroni G, Tsykin A, Lupi M, Gianni M, Goodall GJ
and Garattini E: Induction of miR-21 by retinoic acid in estrogen
receptor-positive breast carcinoma cells: Biological correlates and
molecular targets. J Biol Chem. 286:4027–4042. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Korpal M, Lee ES, Hu G and Kang Y: The
miR-200 family inhibits epithelial-mesenchymal transition and
cancer cell migration by direct targeting of E-cadherin
transcriptional repressors ZEB1 and ZEB2. J Biol Chem.
283:14910–14914. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gregory PA, Bracken CP, Smith E, Bert AG,
Wright JA, Roslan S, Morris M, Wyatt L, Farshid G and Lim YY: An
autocrine TGF-beta/ZEB/miR-200 signaling network regulates
establishment and maintenance of epithelial-mesenchymal transition.
Mol Biol Cell. 22:1686–1698. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Knezevic J, Pfefferle AD, Petrovic I,
Greene SB, Perou CM and Rosen JM: Expression of miR-200c in
claudin-low breast cancer alters stem cell functionality, enhances
chemosensitivity and reduces metastatic potential. Oncogene.
34:5997–6006. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ren Y, Han X, Yu K, Sun S, Zhen L, Li Z
and Wang S: microRNA-200c downregulates XIAP expression to suppress
proliferation and promote apoptosis of triple-negative breast
cancer cells. Mol Med Rep. 10:315–321. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Castilla MÁ, Díaz-Martín J, Sarrió D,
Romero-Pérez L, López-García MÁ, Vieites B, Biscuola M,
Ramiro-Fuentes S, Isacke CM and Palacios J: MicroRNA-200 family
modulation in distinct breast cancer phenotypes. PLoS One.
7:e477092012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen J, Tian W, Cai H, He H and Deng Y:
Down-regulation of microRNA-200c is associated with drug resistance
in human breast cancer. Med Oncol. 29:2527–2534. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Howe EN, Cochrane DR, Cittelly DM and
Richer JK: miR-200c targets a NF-κB up-regulated TrkB/NTF3
autocrine signaling loop to enhance anoikis sensitivity in triple
negative breast cancer. PLoS One. 7:e499872012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Howe EN, Cochrane DR and Richer JK: The
miR-200 and miR-221/222 microRNA families: Opposing effects on
epithelial identity. J Mammary Gland Biol Neoplasia. 17:65–77.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nassirpour R, Mehta PP, Baxi SM and Yin
MJ: miR-221 promotes tumorigenesis in human triple negative breast
cancer cells. PLoS One. 8:e621702013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Søkilde R, Kaczkowski B, Podolska A,
Cirera S, Gorodkin J, Møller S and Litman T: Global microRNA
analysis of the NCI-60 cancer cell panel. Mol Cancer Ther.
10:375–384. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang
Z, Li L, Qin J, Chen Y, Cho WC, et al: miR-221/222 promotes S-phase
entry and cellular migration in control of basal-like breast
cancer. Molecules. 19:7122–7137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Garcia AI, Buisson M, Bertrand P, Rimokh
R, Rouleau E, Lopez BS, Lidereau R, Mikaélian I and Mazoyer S:
Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in
triple negative sporadic breast cancers. EMBO Mol Med. 3:279–290.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kumaraswamy E, Wendt KL, Augustine LA,
Stecklein SR, Sibala EC, Li D, Gunewardena S and Jensen RA: BRCA1
regulation of epidermal growth factor receptor (EGFR) expression in
human breast cancer cells involves microRNA-146a and is critical
for its tumor suppressor function. Oncogene. 34:4333–4346. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Giltnane JM and Balko JM: Rationale for
targeting the Ras/MAPK pathway in triple-negative breast cancer.
Discov Med. 17:275–283. 2014.PubMed/NCBI
|
|
30
|
Hashimoto K, Tsuda H, Koizumi F, Shimizu
C, Yonemori K, Ando M, Kodaira M, Yunokawa M, Fujiwara Y and Tamura
K: Activated PI3K/AKT and MAPK pathways are potential good
prognostic markers in node-positive, triple-negative breast cancer.
Ann Oncol. 25:1973–1979. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lehmann BD, Bauer JA, Chen X, Sanders ME,
Chakravarthy AB, Shyr Y and Pietenpol JA: Identification of human
triple-negative breast cancer subtypes and preclinical models for
selection of targeted therapies. J Clin Invest. 121:2750–2767.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bilir B, Kucuk O and Moreno CS: Wnt
signaling blockage inhibits cell proliferation and migration, and
induces apoptosis in triple-negative breast cancer cells. J Transl
Med. 11:2802013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wahdan-Alaswad R, Harrell JC, Fan Z,
Edgerton SM, Liu B and Thor AD: Metformin attenuates transforming
growth factor beta (TGF-β) mediated oncogenesis in mesenchymal
stem-like/claudin-low triple negative breast cancer. Cell Cycle.
15:1046–1059. 2016. View Article : Google Scholar : PubMed/NCBI
|