Introduction
Gastric cancer (GC) is a frequently diagnosed cancer
among males and females worldwide, and an estimated 951,600 new
stomach cancer cases and 723,100 mortalities occurred in 2012
(1). Although clinical outcome in
therapy of GC has improved unceasingly through earlier diagnosis,
surgical resection, chemotherapy and radiation therapy, the
prognosis of patients with advanced GC remains rather poor with a
five-year survival rate of 5–20% (2,3). The
primary cause of mortality in GC is tumor progression and
metastasis (4). Therefore, the
present study aims to investigate molecular mechanisms that are
involved in the development of GC and provide a novel and effective
method for diagnosis or treatment.
MicroRNAs (miRNAs/miRs) are endogenous small
single-stranded RNA molecules and consist of 18–23 nucleotides. It
has been found that miRNAs are able to participate in the
regulation of cell proliferation, differentiation, metabolism and
apoptosis (5,6). A large number of studies have
subsequently confirmed that miRNAs are involved in the pathogenesis
of a number of human diseases, including cancer (7) and diseases involving the nervous system
(8). In addition, researchers have
also found that miRNAs are able to regulate tumor formation,
infiltration and metastasis by directly binding to the mRNA of
target gene (6,9). Moreover, an increasing number of studies
indicated that miRNAs are involved in the metastasis and invasion
of GC, including miR-34 (10), miR-19
(11), miR-29 (12,13),
miR-495 (14) and miR-551a (14). Although compelling evidence from a
study has indicated that miR-125a-5p was downregulated in GC
(15), the specific mechanisms of
miR-125a-5p in the metastasis and invasion of GC have not been
largely illuminated.
Breast-cancer metastasis suppressor 1 (BRMS1) is a
gene, which inhibits metastasis. First identified in 2000 (16), the gene suppresses the metastasis of
breast carcinoma cells to lungs and regional lymph nodes.
Subsequent studies indicated that BRMS1 mediated inhibition of
metastasis in multiple types of human cancer, including GC
(17–20). A number of studies have confirmed that
a number of protein-coding genes are regulated by miR-125a-5p in
different types of human cancer (21,22).
However, the associations between BRMS1 and miR-125a-5p in GC have
not been fully revealed.
In the present study, a low expression of
miR-125a-5p was frequently observed in GC tissues and cell lines,
and a low miR-125a-5p expression was associated the prognosis of
patients with GC. It has also been demonstrated that miR-125a-5p
expression may affect the invasion and migration of GC cells in
vitro. Based on findings from bioinformatics analysis using
online tools, BRMS1 was identified as a potential target gene of
miR-125a-5p. The experiments demonstrated that miR-125a-5p is able
to regulate the expression of BRMS1 expression in GC cells, which
may lead to further advancements in the knowledge of GC
tumorigenesis.
Materials and methods
Ethics statement
The present study was approved by the Ethics Board
of the Institute of the First Affiliated Hospital of Nanchang
University (Nanchang, China). The ethics board also supervised and
examined the whole process of the present study. All participants
agreed to join the present study and provided written informed
consent.
Gastric tissues samples
Cancer tissue samples and matched normal samples
(distance from tumor, ≥5 cm) were obtained from 82 GC patients (43
males and 39 females; mean age, 58.3 years, age range, 33–85) who
underwent surgical resection from February 2010 to November 2012 at
the Department of General Surgery, First Affiliated Hospital of
Nanchang University (Nanchang, China). The samples were immediately
stored in liquid nitrogen following removal from patients and
detailed clinical data were collected. The histological grade was
assessed according to the tumor-node-metastasis (TNM) system
(23), established by the Union for
International Cancer Control. All patients were monitored every 3
months. The follow-up period ranged from 2 to 80 months (mean
duration, 24.5 months). None of the patients received any
additional treatment prior to surgery.
Cell lines and cell culture
Human GC cell lines (SGC7901, HGC27 and BGC823) were
purchased from the Type Culture Collection of the Chinese Academy
of Sciences (Shanghai, China). The human gastric mucosa GES1 and
HFE145 cell lines were stored in the Central Laboratory of the
Center for Experimental Medicine, the First Affiliated Hospital of
Nanchang University, Nanchang, China), which were obtained from
American Type Culture Collection (Manassas, MA, USA). All cell
lines were cultured in Dulbecco's modified Eagle's medium
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (Invitrogen; Thermo Fisher
Scientific, Inc.) and 100 U/ml penicillin at 37°C with 5%
CO2. The cells in the exponential phrase were used for
experiments.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells or tissues using
a standard Trizol protocol (Invitrogen; Thermo Fisher Scientific,
Inc.). To detect the expression of mRNAs, reverse transcription was
conducted using the GoScriptTM Reverse Transcription system
(Promega Corporation, Madison, WI, USA) according to the
manufacturer's protocol. To detect the level of miRNA expression,
reverse transcription was conducted using the
EnergicScript® cDNA Synthesis kit (ShineGene Molecular
Biotech, Inc., Shanghai, China). Subsequently, the RT-qPCR
detection system FTC-2000 (Funglyn Biotech Inc., Toronto, Canada)
was used to measure the levels of mRNA expression and miRNA with
Shine-SYBR® Real Time qPCR MasterMix kit (ShineGene
Molecular Biotech, Inc.). The expression of mRNA and miRNA was
normalized to endogenous controls β-actin and U6 small nuclear RNA.
Relative fold changes were calculated by the 2−ΔCq
method or the 2−ΔΔCq method (24). The sequences of the primers are listed
as follows: BRMS1 forward, 5′-CAGCCTCCAAGCAAAGACAC-3′ and reverse
5′-GCGGCGTCGCTCATAGTC-3′ and miR-125a-5p forward,
5′-GCTCCCTGAGACCCT-3′ and reverse, 5′-GAGCAGGCTGGAGAA. The
thermocycler conditions were as follows: 94°C for 4 min, then 35
cycles of 94°C for 20 sec, 60°C for 30 sec, and 72°C for 30
sec.
Cell transfection
The human miR-125a-5p plasmids, miR-control
plasmids, anti-miR-125a-5p and anti-miR-control were designed and
generated by Shanghai GenePharma Co., Ltd., Shanghai, China
(Shanghai, China). The siRNA small-interfering (si)RNA (#S1
sequence, 5′-GGAAUAAGUACGGAAUGUGA-3′) that targets the BRMS1 gene
was synthesized by Guangzhou RiboBio Co., Ltd., (Guangzhou, China)
as previously described (25).
Additionally, the coding sequences [without the 5′-untranslated
region (UTR) of BRMS1] was amplified by PCR and cloned into the
pcDNA3.1 (+) vector (Shanghai GenePharma Co., Ltd). The
thermocycler conditions were as follows: 94°C for 5 min, 94°C for 1
min and 50°C for 1 min, followed by 5 cycles of 72°C for 1 min,
followed by 25 cycles of 94°C for 1 min, 65°C for 1 min and of 72°C
for 1 min, All constructs were verified by sequencing. The
miR-125a-5p plasmids, miR-control plasmids, anti-miR-125a-5p and
anti-miR-control (40 nM) were transfected using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) into cells in 12-well plates when 80–90%
confluence was reached. A total of 24 h following transfection, the
cells were used to perform migration and invasion assays. The RNA
analysis and protein analysis were performed at 48 h following
transfection. All sequences and primers used in this section were
listed in Table I.
 | Table I.All primers and sequences used in the
present study. |
Table I.
All primers and sequences used in the
present study.
| Name | Sequence
(5′-3′) |
|---|
| qPCR |
|
| BRMS1
Forward |
CAGCCTCCAAGCAAAGACAC |
| BRMS1
Reverse |
GCGGCGTCGCTCATAGTC |
|
miR-125a-5p Forward |
GCTCCCTGAGACCCT |
|
miR-125a-5p Reverse |
GAGCAGGCTGGAGAA |
| β-actin
Forward |
TGACGTGGACATCCGCAAAG |
| β-actin
Reverse |
CTGGAAGGTGGACAGCGAGG |
| U6
snRNA Forward |
CTCGCTTCGGCAGCACA |
| U6
snRNA Reverse |
AACGCTTCACGAATTTGCGT |
| Luciferase
assays |
|
|
BRMS15′UTR(WT) Forward |
CCGCTCGAGAAGCACCGATAGGCTCTGCCTC |
| BRMS1
5′UTR (WT) Reverse |
CGGGATCCCTGGACTCGCGGGGACTGG |
| BRMS1
5′UTR(MU) Forward |
CCGCTCGAGAAGCACCGATAGGCTCTGCCTC |
| BRMS1
5′UTR (WT) Reverse |
CGGGATCCCTGGCCTCGCGGGGACTGGAGCCTCTGGCCTCACGACGGAGATTGGGACTCAGCTGCCC |
| miRNAs and
siRNAs |
|
| BRMS1
Forward |
CCGCTCGAGGCCACCATGCCTGTCCAGCCTCCAAG |
| BRMS1
Reverse |
CGCGGGCCCTCACTTGTCGTCATCGTCCTGTAGTCAGGTCCATCCGATTTTCTCTTCT |
|
Si-BRMS1 Forward |
GATCCGGAATAAGTACGAATGTGATTCAAGAGATCACATTCGTACTTATTCTTTTTTG |
|
Si-BRMS1 Reverse |
AATTCAAAAAAGGAATAAGTACGAATGTGATCTCTTGAATCACATTCGTACTTATTCCG |
|
miR-125a-5p sense |
GAGCUCUCCCUGAGACCCUUUAACCUGUGAAAGCUU |
|
miR-125a-5p antisense |
AAGCUUUCACAGGUUAAAGGGUCUCAGGGAGAGCUC |
|
miR-125a-5p control sense |
UUCUCCGAACGUGUCACGUTT |
|
miR-125a-5p control
antisense |
ACGUGACACGUUCGGAGAATT |
Cell migration and invasion
analysis
The invasive and migratory capacity of the cells was
estimated using Transwell chambers (diameter, 6.5 mm, membrane pore
size, 8 µm; Corning Incorporated, Corning, NY, USA). To conduct
invasion assays, the membranes were coated with 1 mg/ml Matrigel
(BD Biosciences, Franklin Lakes, NJ USA), Matrigel was not used for
migration assays. The cells (migration assay, 5×104
cells; invasion assay, 1×105 cells) were suspended in
200 µl serum-free medium, and the cells were added to the upper
chamber. Then, 600 µl 20% FBS-DMEM was added to the lower chamber.
Following incubation at 37°C for 24 h, the non-migrating or
non-invading cells were removed with cotton swabs. Finally, invaded
cells on the lower side of the filter were fixed with 4%
paraformaldehyde for 15 min and stained with 0.1% crystal violet
(Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) at 37°C for 30 min.
The cells were counted in five different fields with a microscope
(Olympus Corporation, Tokyo, Japan).
Bioinformatics analysis
MiR-125a-5p target genes were identified using four
web-based bioinformatics algorithms: microRNA.org
(http://www.microrna.org/microrna/home.do), MicroCosm
Targets (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/info.html),
TargetScan Human (http://www.targetscan.org/vert_50/) and miRTar
(http://mirtar.mbc.nctu.edu.tw/human/), which predict
miRNA-binding sites based on complementarity to the nucleotide
sequence of the miRNA (all accessed on November 10th, 2014). The
algorithms used identified highly complementary sites. The results
of miRTar indicated that the 5′UTR of BRMS1 binds to miR-125a-5p
with a high score.
Luciferase assays
The 5′-UTR of the BRMS1 segment was amplified by PCR
and inserted into the pHY-LV-Report 3.1 vector (http://www.hanyinbt.com; Shanghai, China). The
thermocycler conditions were as follows: 94°C for 2 min, followed
by 35 cycles at 94°C for 20 sec, 68°C for 40 sec and 72°C for 2 min
The mutant of the seed region of the putative miR-125a-5p binding
sites in the BRMS1 5′UTR was generated using a QuikChange
Site-Directed Mutagenesis kit (Agilent Technologies, Inc., Santa
Clara, CA, USA). HGC27 cells (8×103 cells) were seeded
into the 96-well plates 24 h prior to transfection. A mixture of
pHY-LV-5′-UTR, negative miR-control (miR-NC) or 40 nM miR-125a-5p
plasmids were co-transfected with Renilla into HGC27 cells
using Lipofectamine® 2000 reagent (Invitrogen, Thermo
Fisher Scientific, Inc.). 24 h later, the luciferase activity was
measured using the Dual Luciferase assay (Promega Corporation). The
Renilla reporter vector was used as an internal control to
assess the efficiency of transfection. The primer sequences were
listed in Table I.
Western blot assays
Total protein was extracted from gastric tissues and
cells by Total Protein Extraction kit (Nanjing KeyGen Biotech Co.,
Ltd., Nanjing, China) according to the manufacturer's protocol and
using the BAC kit (Tiangen Biotech Co., Ltd., Beijing, China) to
detect the concentration of the proteins. The proteins were
separated by 12% SDS-PAGE and transferred onto polyvinyl fluoride
(PVDF) membranes (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
The membranes were blocked with 5% non-fat dried milk for 1 h at
room temperature. Then, the membranes were incubated with the
primary monoclonal antibody against BRMS1 (1:500; Sigma-Aldrich;
Merck KGaA; catalog no. WH0025855M1, Merck KGaA) or β-actin
(1:1,000; Cell Signaling Technology, Inc., catalog no. 8H10D10)
overnight at 4°C. After washing the membranes with TBST (TBS with
0.1% Tween-20) three times, the membranes were incubated for 2 h at
room temperature, with horseradish peroxidase-conjugated secondary
antibody (goat anti-mouse IgG; Abcam, Cambridge, UK; catalog no.
ab97023) and the ECL Western Blotting Analysis system (GE
Healthcare, Chicago, IL, USA) was used to detect the levels of
expression of the target proteins. Band intensities were quantified
using Image-Pro Plus software (version, 6.0; Media Cybernetics,
Inc., Rockville, MD, USA).
Statistical analysis
All experiments in the present study were repeated
at least three times. The data are presented as the mean ± standard
deviation and P<0.05 was considered to indicate a statistically
significant difference. SPSS (version, 19.0; IBM Corp., Armonk, NY,
USA) software was used for statistical analysis. Differences
between the groups were estimated using the χ2,
Student's t-test and one-way analysis of variance with a
Student-Newman-Keuls post-hoc test. Survival was evaluated using
the Kaplan-Meier method, and the correlation between miR-125a-5p
and BRMS1 protein expression level was evaluated using Pearson's
correlation.
Results
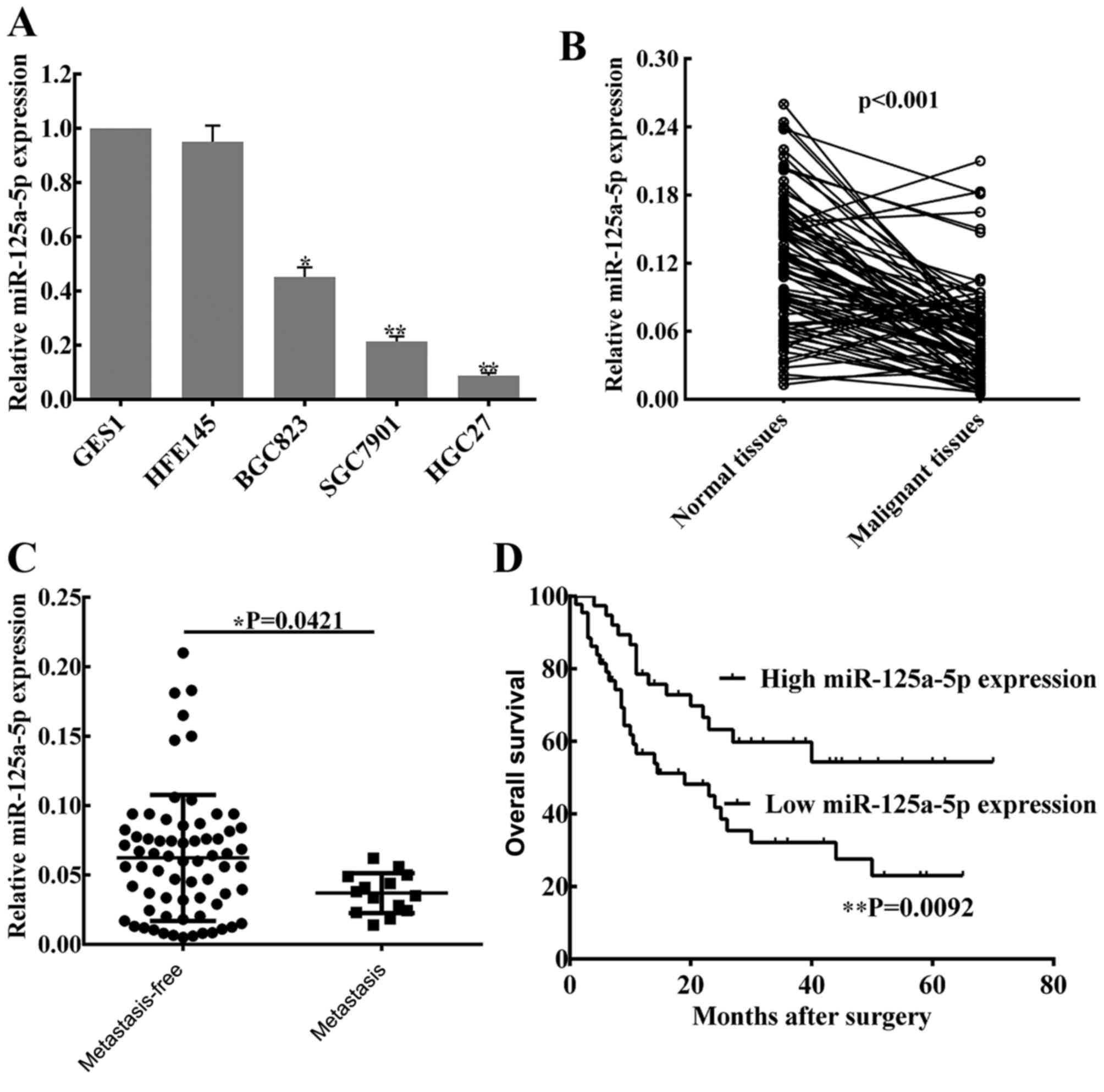
miR-125a-5p is downregulated in GC
cells and tissues
To determine the level of miR-125a-5p expression in
GC, three malignant human GC cell lines (SGC7901, HGC27 and BGC823)
and two normal gastric mucosa cell lines (GES1 and HFE145), as well
as 82 pairs of cancer tissues and matched normal tissues from
patients with GC were used to perform RT-qPCR analysis. It was
observed that the levels of miR-125a-5p were significantly lower in
the GC cell lines compared with the expression in normal gastric
mucosa cell lines (Fig. 1A), whereas
no statistical difference in miR-125a-5p expression was indicated
between the two normal gastric mucosa cell lines. In patient
tissues, miR-125a-5p expression was lower compared with the matched
normal tissues (P<0.01; Fig. 1B).
In addition, based on clinical progression, the expression of
miR-125a-5p was markedly decreased in patients with peritoneal
metastasis compared with patients without peritoneal metastasis
(P=0.0421; Fig. 1C).
Levels of miR-125a-5p are associated
with clinical pathological characteristics and prognosis in
patients with GC
To investigate the associations between miR-125a-5p
expression and clinical pathological characteristics, the data of
82 patients was collected from the Pathology Department of the
First Affiliated Hospital of Nanchang University, and the detailed
information is listed in Table II.
The results showed that the expression of miR-125a-5p was
significantly associated with lymph node metastasis (P=0.034),
peritoneal dissemination (P=0.030) and advanced TNM stage
(P=0.025). However, no associations were identified between
miR-125a-5p expression and other clinical pathological
characteristics. Notably, Kaplan-Meier survival curves of patients
with GC indicate that the overall survival rates of GC patients
with low expression of miR-125a-5p was significantly shorter
compared with patients with high miR-125a-5p expression (P=0.0092;
Fig. 1D).
 | Table II.Clinicopathological characteristics
of patients with gastric cancer and miR-125a-5p expression in tumor
tissues. |
Table II.
Clinicopathological characteristics
of patients with gastric cancer and miR-125a-5p expression in tumor
tissues.
|
|
| miR-125a-5p
expression (n, %) |
|
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics | n | Low | High | X2 | P-value |
|---|
| Age, years |
|
|
|
|
|
|
<65 | 58 | 37 (72.5) | 21 (67.7) | 0.215 | 0.643 |
|
≥65 | 24 | 14 (27.5) | 10 (32.3) |
|
|
| Sex |
|
|
|
|
|
|
Male | 43 | 24 (47.1) | 19 (61.3) | 1.566 | 0.211 |
|
Female | 39 | 27 (52.9) | 12 (38.7) |
|
|
| Tumor size, cm |
|
|
|
|
|
|
>3.5 | 27 | 19 (32.8) | 8 (33.3) | 0.003 | 0.960 |
|
≤3.5 | 55 | 39 (67.2) | 16 (66.7) |
|
|
| Tumor location |
|
|
|
|
|
|
Proximal | 17 | 12 (19.4) | 5 (25.0) |
|
|
|
Middle | 18 | 14 (22.6) | 4 (20.0) | 0.303 | 0.859 |
|
Distal | 47 | 36 (58.0) | 11 (55.0) |
|
|
|
Differentiation |
|
|
|
|
|
|
Well | 15 | 14 (25.0) | 1 (6.2) |
|
|
|
Moderate | 26 | 22 (39.4) | 4 (25.0) | 3.256 | 0.196 |
|
Poor | 41 | 30 (53.6) | 11 (68.8) |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
Absent | 23 | 13 (21.7) | 10 (45.5) | 4.514 | 0.034a |
|
Present | 59 | 47 (78.3) | 12 (54.5) |
|
|
| Peritoneal
metastasis |
|
|
|
|
|
|
Absent | 68 | 43 (76.8) | 25 (96.2) | 4.705 | 0.030a |
|
Present | 14 | 13 (23.2) | 1 (3.8) |
|
|
| TNM stage |
|
|
|
|
|
|
I–II | 40 | 21 (39.6) | 19 (65.5) | 5.030 | 0.025a |
|
III–IV | 42 | 32 (60.4) | 10 (34.5) |
|
|
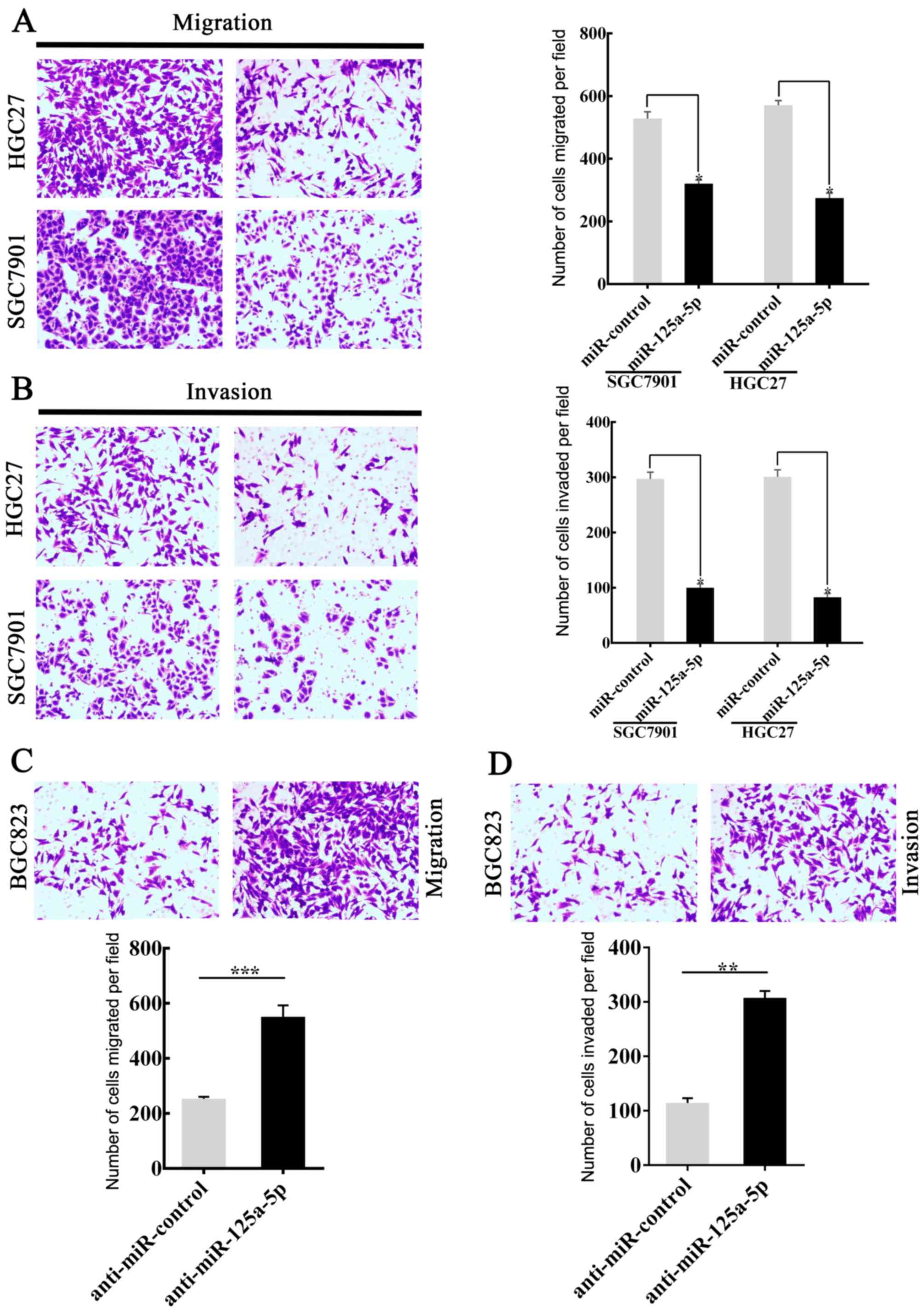
miR-125a-5p regulates the invasion and
migration of GC cells in vitro
Previously, the association between miR-125a-5p and
GC metastasis was observed (as aforementioned) and the expression
of miR-125a-5p was downregulated in patients with gastric cancer,
it was hypothesized that a lower miR-125a-5p expression in GC
tissues was involved in invasion and metastasis of GC cells. In
order to investigate the hypothesis, SGC7901 and HGC27 cells were
successfully transfected with miR-125a-5p to generate an
miR-125a-5p-overexpression model. The miR-control was also
transfected. Invasion and migration analyses were then performed.
The results showed that the migratory (P<0.05; Fig. 2A) and invasive (P<0.05; Fig. 2B) capacity of SGC7901 and HGC27 cells
transfected with miR-125a-5p were significantly lower compared with
those of the control group. By contrast, BGC823 cells, with high
levels of endogenous miR-125a-5p expression, were transfected with
anti-miR-125a-5p. Migration and invasion analyses were subsequently
performed. The migration (P<0.001; Fig. 2C) and invasion (P<0.01; Fig. 2D) analyses indicated that the
downregulation of miR-125a-5p was able to accelerate the movement
of BGC823 cells from the upper chamber to the lower chamber. The
level of miR-125a-5p expression in GC cells following transfection
is shown in Fig. 1. Taken together,
the data was able to confirm the hypothesis that the level of
miR-125a-5p expression affects the migratory and invasive abilities
of GC cells.
BRMS1 is a direct target of
miR-125a-5p
It has been verified that miRNAs generally regulate
the expression of target genes to regulate cellular processes
associated with cancer, including metastasis (26). Therefore, bioinformatics websites,
including microRNA.org, MicroCosm Targets,
TargetScan Human and miRTar were used to predict whether
miR-125a-5p target BRMS1 genes. Bioinformatics analysis indicated
that there is a putative binding site for miR-125a-5p in the 5′-UTR
of BRMS1 mRNA (Fig. 3A). To
investigate whether BRMS1 is an exact target of miR-125a-5p, the
full-length wild-type (Wt-BRMS1-5′UTR) and mutant BRMS1 5′-UTRs
(Mt-BRMS1-5′UTR) were amplified and directly fused to the
pHY-LV-Report 3.1 vector downstream of the luciferase reporter gene
(Fig. 3A). Then, luciferase assays
were performed, and HGC27 cells were co-transfected with the vector
and either with miR-NC or miR-125a-5p. Luciferase activity was
significantly decreased in the group that transfected with
miR-125a-5p and Wt-BRMS1-5′UTR compared with the other three groups
(P<0.05; Fig. 3B).
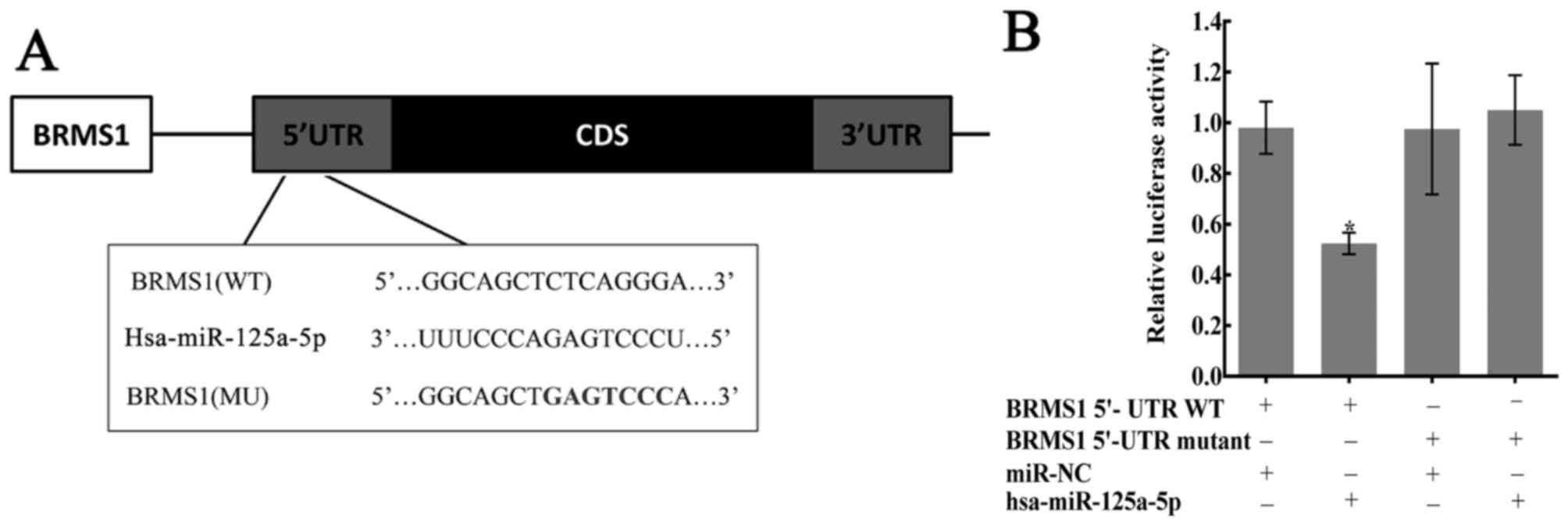 | Figure 3.Confirmation of predicted binding
sites between miR-125a-5p and BRMS1. (A) The bioinformatics
websites (microRNA.org, MicroCosm Targets,
TargetScan Human and miRTar) predicted that the 5′-UTR of BRMS1
mRNA contains the binding sequences of miR-125a-5p. (B) Dual
luciferase reporter assay performed in HGC27 cells. The average
values of normalized 5′-UTR luciferase intensity were calculated
from three independent experiments. Luciferase activity was
significantly decreased in the group transfected with miR-125a-5p
and Wt-BRMS1-5′UTR, compared with the other three groups,
*P<0.05. BRMS1, breast cancer metastasis suppressor 1; CD,
coding DNA sequence; miR, microRNA; MU, mutated; UTR, untranslated
region; WT, wild-type. |
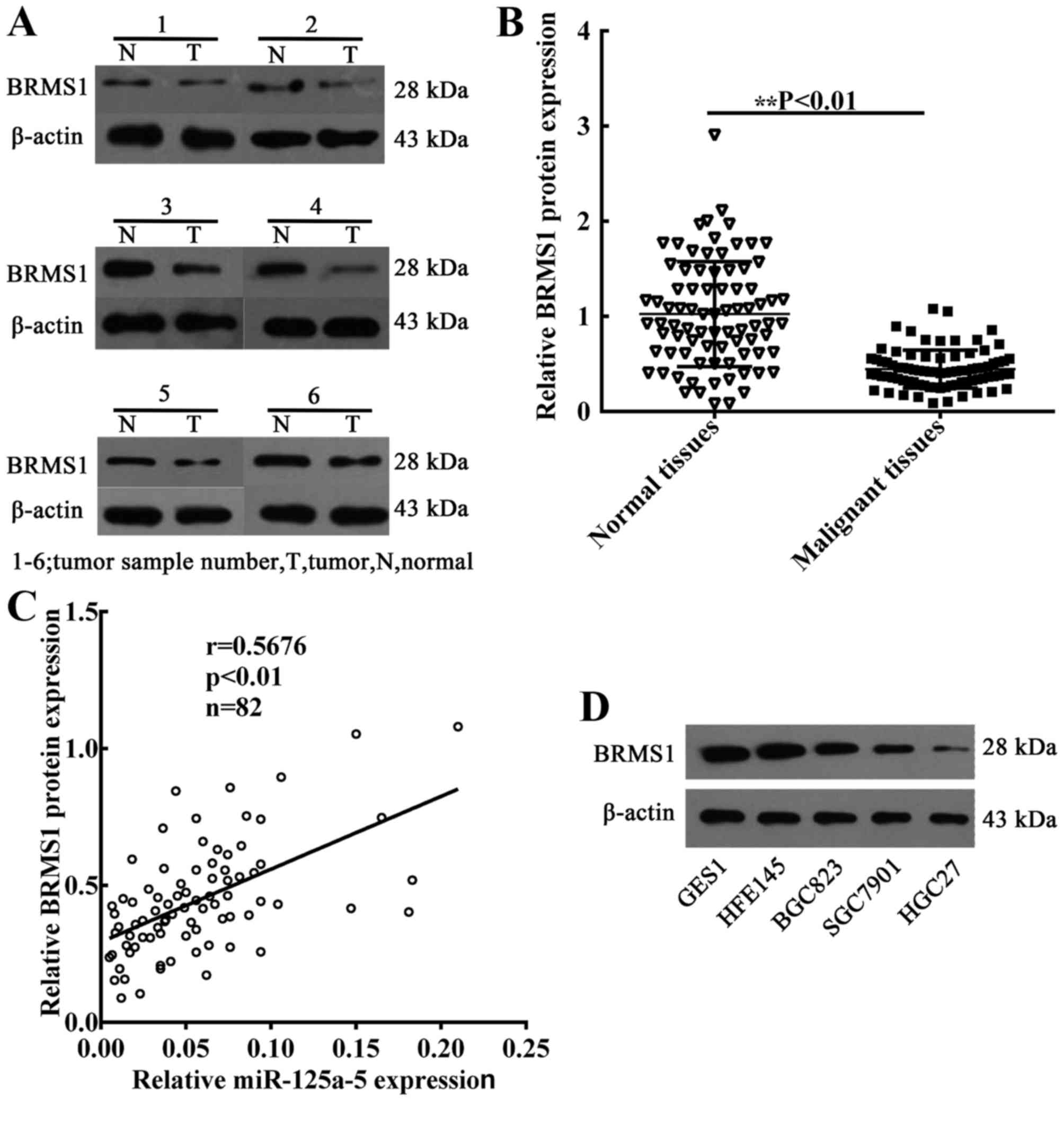
miR-125a-5p expression is positively
correlated with the levels of BRMS1 in GC
To determine the association between miR-125a-5p
expression and BRMS1 protein levels, the expression of BRMS1 in 82
GC patient tissues was also detected. The mean levels of BRMS1
protein were also decreased in GC samples compared with the
expression in normal gastric samples (Fig. 4A and B). Subsequently, linear
correlation analysis was performed to analyze the correlation
between BRMS1 protein expression and the levels of miR-125a-5p in
GC samples. The findings revealed that the level of BRMS1 protein
was positively correlated with the levels of miR-125a-5p
(P<0.01; Fig. 4C). In addition, in
order to verify that miR-125a-5p is able to regulate BRMS1 protein
expression in GC cells, western blot assays were performed to
detect the levels of BRMS1 protein in GC cell lines. Compared with
GES1 and HFE145 cells, the expression of BRMS1 protein was
significantly decreased in GC cells (Fig.
4D).
miR-125a-5p regulates BRMS1 protein
expression in GC cell lines
To further verify the effects of miR-125a-5p on the
regulation of BRMS1 expression, RT-qPCR and western blotting were
performed to detect the relative mRNA and protein expression level
in various GC cell lines (SGC7901, HGC27, BGC823). The results
indicated that overexpression of miR-125a-5p (Fig. 5A) increased BRMS1 mRNA and protein
expression in SGC7901 (Fig. 5B) and
HGC27 cells (Fig. 5C). By contrast,
marked suppression of BRMS1 expression was observed in
inhibitor-treated BGC823 cells compared with untreated cells and
negative control-treated cells (Fig.
5D).
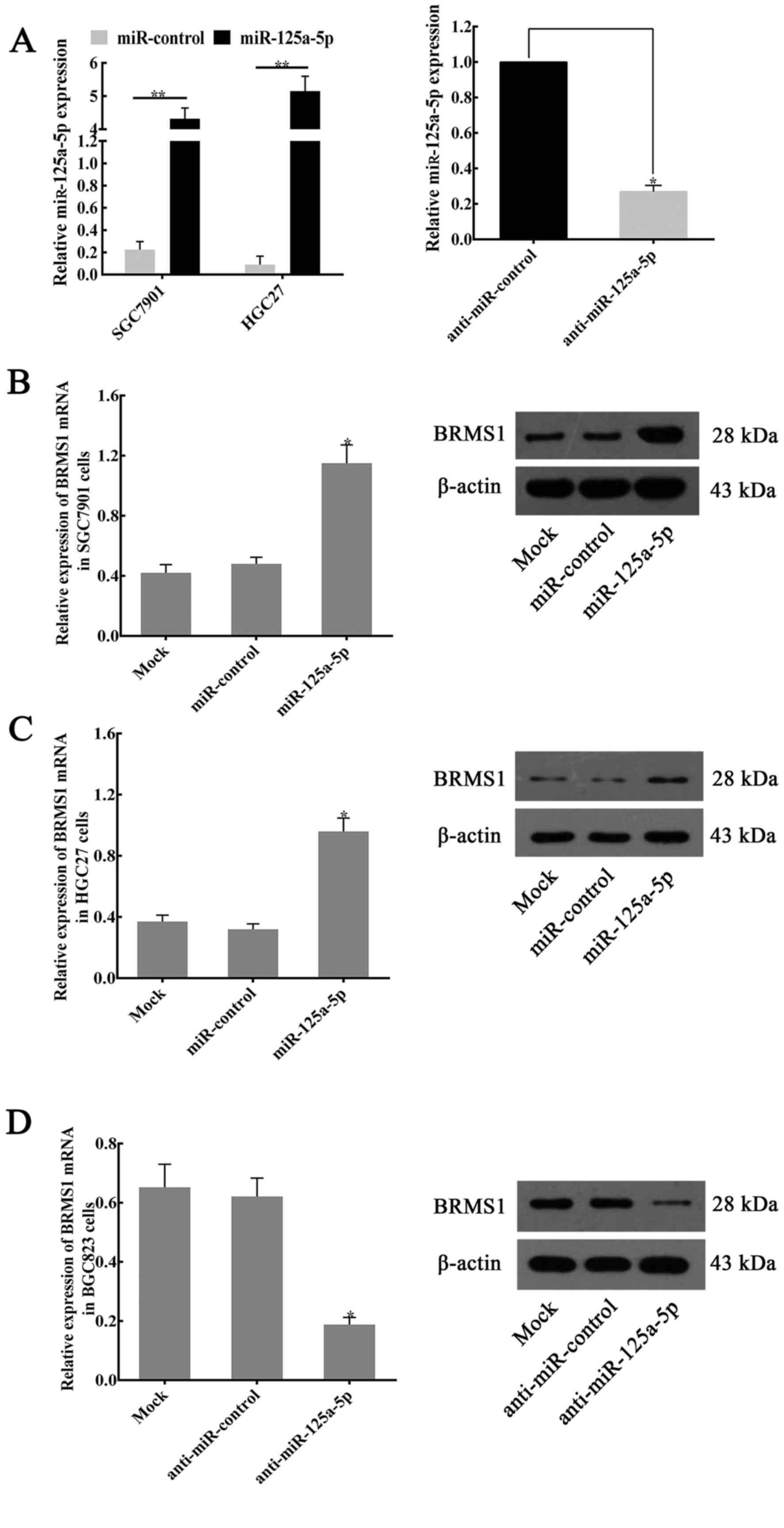 | Figure 5.miR-125a-5p regulates the expression
of BRMS1 in GC cell lines. (A) Compared with the expression of
miR-125a-5p in miR-control-transfected controls, miR-125a-5p
expression in miR-125a-5p plasmid-transfected cells was
upregulated. By contrast, following the transfection of
anti-miR-125a-5p, the miR-125a-5p expression in BGC823 cells was
significantly suppressed. *P<0.05, **P<0.01. The expression
of BRMS1 mRNA and protein in (B) SGC7901 and (C) HGC27 cells
following the transfection of miR-NC or miR-125a-5p. *P<0.05 vs.
Mock or miR-control groups. (D) Following transfection of BGC823
cells with anti-miR-125a-5p, the expression of BRMS1 mRNA and
protein was significantly decreased compared with the expression in
anti-miR-NC or mock groups. *P<0.05. For reverse
transcription-quantitative polymerase chain reaction assays,
β-actin was used as an internal control for expression of BRMS1,
and the values indicate the mean ± standard deviation (n=3). For
western blotting, β-actin served as an internal control. BRMS1,
breast cancer metastasis suppressor 1; miR, microRNA; NC, negative
control; si, small-interfering. |
miR-125a-5p inhibits migration and
invasion of gastric cancer cells via BRMS1
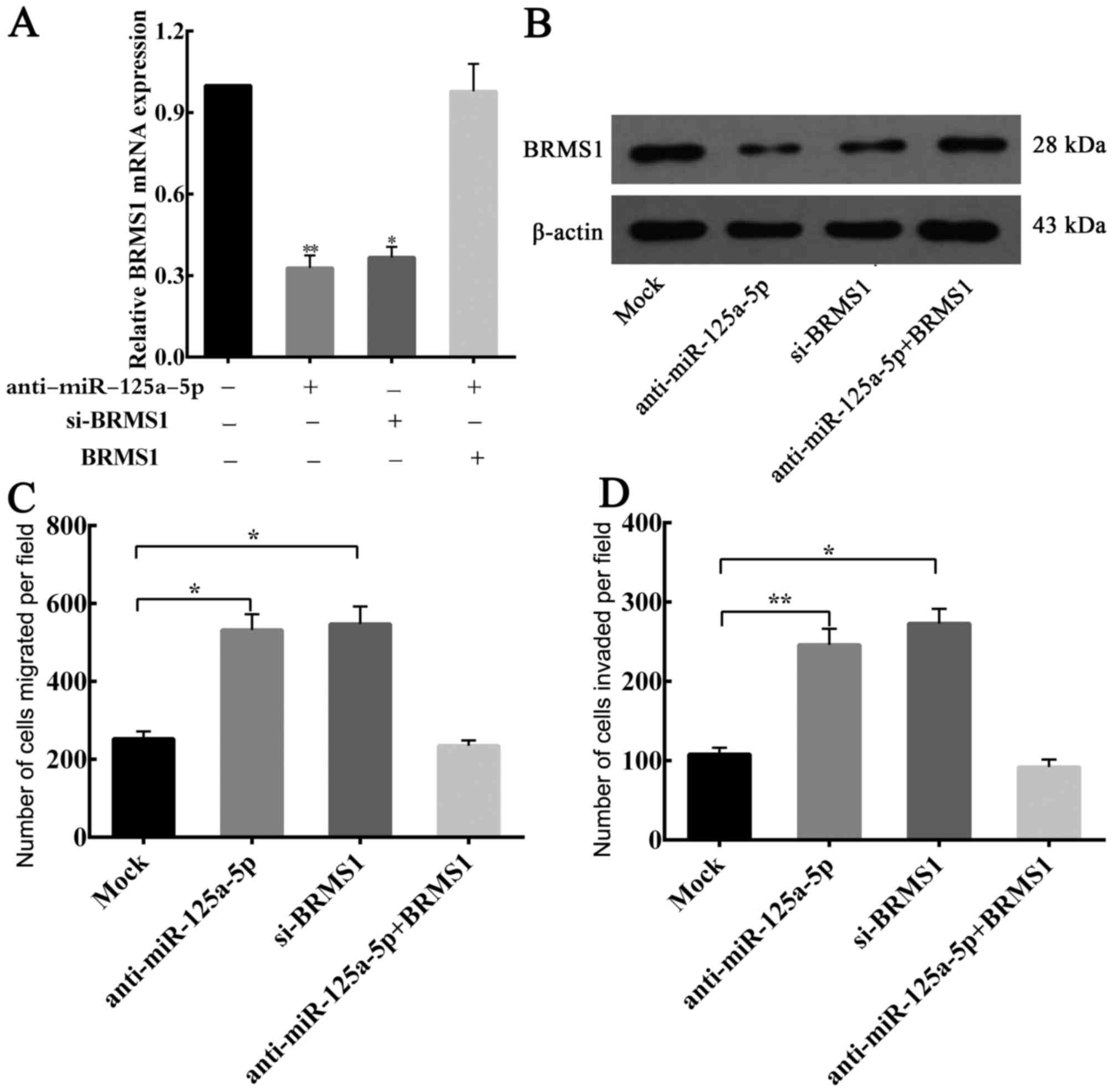
Considering the aforementioned results, whether
BRMS1 is a functional target of endogenous miR-125a-5p, which
affects GC cells migration and invasion, was investigated. BGC823
cells were transfected separately with si-BRMS1 and
anti-miR-125a-5p, and BRMS1 expression was analyzed by RT-qPCR and
western blotting. The transfection efficiency of si-BRMS1 and
anti-miR-125a-5p was detected (Fig. 6A
and B). Furthermore, it was observed that there was no
statistical significant difference in the ability to downregulate
endogenous BRMS1 expression between anti-miR-125a-5p and
si-BRMS1.
Migration and invasion assays revealed that
knockdown of BRMS1 mRNA was able to significantly increase the
migration and invasion of the BGC823 cells, and similar results
were observed when there is a low expression of miR-125a-5p in GC
cells (Fig. 6C and D). Moreover, when
BRMS1 expression was restored, it was able to counteract the
inhibitory effects of anti-miR-125a-5p in BGC823 cells (Fig. 6A-D). Taken together, these results
confirmed that miR-125a-5p is able to affect the migration and
invasion of GC cells through regulating BRMS1 expression.
Discussion
In the present study, it was observed that
miR-125a-5p is able to act as a tumor suppressor in GC. It was also
revealed that downregulation of miR-125a-5p was a risk factor for
lymph node and peritoneal metastasis in patients with GC.
In GC cells, upregulated miR-125a-5p expression was
able to inhibit cell invasion and migration. Furthermore, to the
best of our knowledge, BRMS1 was identified as a potential novel
target of miR-125a-5p, and BRMS1 was associated with peritoneal
metastasis in GC.
It has been hypothesized that miRNAs may function as
tumor suppressors or tumor promoters, and thus perform critical
roles in tumor development and progression (9,27).
miR-125a-5p is located at 19q13.41, and its downregulated
expression in non-small cell lung cancer (NSCLC) was first observed
in 2006 (28). A number of studies
have demonstrated that the role of miR-125a-5p as a
tumor-inhibiting factor in several types of tumors, including
breast cancer, NSCLC and ovarian cancer (29–31).
Additionally, Tong et al (32)
have demonstrated that miR-125a-5p miRNA was downregulated in
colorectal cancer. From these data, it may be inferred that
miR-125a-5p act as a potential tumor suppressor in tumors involving
the digestive tract. However, the association between miR-125a-5p
and GC has not been widely investigated. In the present study, it
was observed that there was a low expression of miR-125a-5p in
primary tumor tissues obtained from patients with GC compared with
matched normal tissues.
In addition, miR-125a-5p expression was negatively
associated with lymph node metastasis, peritoneal dissemination and
advanced TNM stage. In vitro experiments also indicated that
overexpression of miR-125a-5p was able to suppress the migration
and invasion of GC cells.
As part of the present study on how the loss of
miR-125a-5p affects GC metastasis, it was confirmed that BRMS1 was
a critical downstream target of miR-125a-5p. BRMS1 is a tumor
suppressor (33). It has been
verified that the expression of BRMS1 was regulated by miRNAs, and
that it is able to inhibit the process of epithelial-mesenchymal
transition and invasion in breast cancer (33). BRMS1 also regulates a network of
proteins with central roles in cancer metastasis (19,34). For
instance, Mei et al (35)
demonstrated that BRMS1 was able to inhibit the invasion of glioma
cells by suppressing urokinase-type plasminogen activator, nuclear
factor-κB and the expression and enzymatic activity of matrix
metalloproteinase-2 (MMP-2). You et al (36) reported that BRMS1 is able to regulate
apoptosis in NSCLC cells by modulating the activation of signal
transducer and activator of transcription 3. Apart from the
aforementioned signaling pathways, decreased expression of BRMS1
has also been demonstrated to induce the upregulation of human
epidermal growth factor receptor 2 (HER2) in breast cancer
(37). The deregulation of HER2 in
cancer has been increasingly recognized, and HER2 may be an
important therapeutic target in GC. A further study revealed that
overexpression of miR-125a-5p was able to inhibit invasion and
metastasis in GC by regulating gene HER2 (15). Based on these findings, it is
hypothesized that miR-125a-5p and BRMS1 have pivotal implications
in GC.
To further study the regulatory mechanisms between
miR-125a-5p and BRMS1, target prediction programs were used to
predict BRMS1 mRNA binding site that binds with miR-125a-5p. It was
identified that the 5′-UTR of BRMS1 contained a conserved putative
target site for miR-125a-5p.
In previous studies, miRNAs have been frequently
reported to negatively regulate gene expression and pair with the
3′-UTR of specific target mRNAs (12–14,38–40).
A number of studies have indicated that miRNAs are able to
upregulate the translation of the target mRNAs and promote protein
expression, by directly interacting with the 5′UTR of the mRNA
(41–43). In the present study, luciferase
reporter assays revealed that miR-125a-5p is able to directly
interact with the 5′UTR of BRMS1. Exogenous expression of
miR-125a-5p resulted in increased BRMS1 expression and inhibited
the invasion and metastasis of GC cells. By contrast, the knockdown
of miR-125a-5p induced the loss of BRMS1 expression, and
upregulated the invasion and metastasis of GC cells. Restoration of
BRMS1 expression resulted in the suppression of invasion and
migration of GC cells. To the best of our knowledge, the results
from the present study is the first to indicate that miR-125a-5p is
able to directly target and promote BRMS1 expression by binding to
the 5′UTR of BRMS1 mRNA.
In summary, miR-125a-5p was identified as a
potential tumor suppressor in GC. Additionally, miR-125a-5p
inhibits the metastatic characteristics of GC cells in
vitro, including migration and invasion. Moreover, BRMS1 was
identified as a potential target gene of miR-125a-5p in the
development of GC. Therefore, further investigation of the
miR-125a-5p-BRMS1 axis may provide novel therapeutic strategies for
the treatment of GC. Further study is required to fully elucidate
the mechanism of the miR-125a-5p-BRMS1 axis.
Acknowledgements
The authors thank the Departments of General Surgery
and Pathology at the First Affiliated Hospital of Nanchang
University for providing tissue samples and related clinical
data.
Funding
The present study was supported by the National
Science Foundation of China (grant nos. 81460373 and 81360362) and
the ‘Talent 555 Project’ of Jiangxi, China.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZGJ conceived the study; ZGJ, ZRL, YC, SXT and YT
designed the experiments; SXT, GYZ, YL, DJL, SX and JBX identified,
designed, or performed the methods; SXT, GYZ, DJL, ZBL, WYZ and PTG
performed the experiments; SXT, GYZ and YC analyzed the data; and
YC, SXT and ZGJ wrote the paper.
Ethics approval and consent to
participate
The present study was approved by the Ethics Board
of the Institute of the First Affiliated Hospital of Nanchang
University (Nanchang, China). The ethics board also supervised and
examined the whole process of the present study. All participants
agreed to join the present study and provided written informed
consent.
Consent for publication
All participants provided written informed consent
for publication of the present study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yasui W, Sentani K, Sakamoto N, Anami K,
Naito Y and Oue N: Molecular pathology of gastric cancer: Research
and practice. Pathol Res Pract. 207:608–612. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hartgrink HH, Jansen EP, van Grieken NC
and van de Velde CJ: Gastric cancer. Lancet. 374:477–490. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen DL, Zhang DS, Lu YX, Chen LZ, Zeng
ZL, He MM, Wang FH, Li YH, Zhang HZ, Pelicano H, et al:
microRNA-217 inhibits tumor progression and metastasis by
downregulating EZH2 and predicts favorable prognosis in gastric
cancer. Oncotarget. 6:10868–10879. 2015.PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiao X, Tang C, Xiao S, Fu C and Yu P:
Enhancement of proliferation and invasion by MicroRNA-590-5p via
targeting PBRM1 in clear cell renal carcinoma cells. Oncol Res.
20:537–544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fernandez-Santiago R, Iranzo A, Gaig C,
Serradell M, Fernández M, Tolosa E, Santamaría J and Ezquerra M:
MicroRNA association with synucleinopathy conversion in rapid eye
movement behavior disorder. Ann Neurol. 77:895–901. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: Oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang AM, Huang TT, Hsu KW, Huang KH, Fang
WL, Yang MH, Lo SS, Chi CW, Lin JJ and Yeh TS: Yin Yang 1 is a
target of microRNA-34 family and contributes to gastric
carcinogenesis. Oncotarget. 5:5002–5016. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu Q, Yang Z, An Y, Hu H, Yin J, Zhang P,
Nie Y, Wu K, Shi Y and Fan D: MiR-19a/b modulate the metastasis of
gastric cancer cells by targeting the tumour suppressor MXD1. Cell
Death Dis. 5:e11442014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han TS, Hur K, Xu G, Choi B, Okugawa Y,
Toiyama Y, Oshima H, Oshima M, Lee HJ, Kim VN, et al: MicroRNA-29c
mediates initiation of gastric carcinogenesis by directly targeting
ITGB1. Gut. 64:203–214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gong J, Li J, Wang Y, Liu C, Jia H, Jiang
C, Wang Y, Luo M, Zhao H, Dong L, et al: Characterization of
microRNA-29 family expression and investigation of their
mechanistic roles in gastric cancer. Carcinogenesis. 35:497–506.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Z, Cao Y, Jie Z, Liu Y, Li Y, Li J, Zhu
G, Liu Z, Tu Y, Peng G, et al: miR-495 and miR-551a inhibit the
migration and invasion of human gastric cancer cells by directly
interacting with PRL-3. Cancer Lett. 323:41–47. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nishida N, Mimori K, Fabbri M, Yokobori T,
Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y and Mori M:
MicroRNA-125a-5p is an independent prognostic factor in gastric
cancer and inhibits the proliferation of human gastric cancer cells
in combination with trastuzumab. Clin Cancer Res. 17:2725–2733.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Seraj MJ, Samant RS, Verderame MF and
Welch DR: Functional evidence for a novel human breast carcinoma
metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer
Res. 60:2764–2769. 2000.PubMed/NCBI
|
|
17
|
Wu J, Wang Y, Qiao X, Saiyin H, Zhao S,
Qiao S and Wu Y: Cloning and characterization of a novel human
BRMS1 transcript variant in hepatocellular carcinoma cells. Cancer
Lett. 337:266–275. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu Y, Mayo MW, Nagji AS, Hall EH, Shock
LS, Xiao A, Stelow EB and Jones DR: BRMS1 suppresses lung cancer
metastases through an E3 ligase function on histone
acetyltransferase p300. Cancer Res. 73:1308–1317. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Slipicevic A, Holm R, Emilsen E, Ree
Rosnes AK, Welch DR, Mælandsmo GM and Flørenes VA: Cytoplasmic
BRMS1 expression in malignant melanoma is associated with increased
disease-free survival. BMC Cancer. 12:732012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Guan J, Sun Y, Chai J, Zou T,
Gong W, Zhu Z, Liu X, Hou Q and Song X: Effect of BRMS1 on
tumorigenicity and metastasis of human rectal cancer. Cell Biochem
Biophys. 70:505–509. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hsieh TH, Hsu CY, Tsai CF, Long CY, Chai
CY, Hou MF, Lee JN, Wu DC, Wang SC and Tsai EM: miR-125a-5p is a
prognostic biomarker that targets HDAC4 to suppress breast
tumorigenesis. Oncotarget. 6:494–509. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jiang L, Huang Q, Chang J, Wang E and Qiu
X: MicroRNA HSA-miR-125a-5p induces apoptosis by activating p53 in
lung cancer cells. Exp Lung Res. 37:387–398. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Washington K: 7th edition of the AJCC
cancer staging manual: Stomach. Ann Surg Oncol. 12:3077–3079. 2010.
View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)). Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu Y, Jiang W, Wang Y, Wu J, Saiyin H,
Qiao X, Mei X, Guo B, Fang X, Zhang L, et al: Breast cancer
metastasis suppressor 1 regulates hepatocellular carcinoma cell
apoptosis via suppressing osteopontin expression. PLoS One.
7:e429762012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Garofalo M and Croce CM: microRNAs: Master
regulators as potential therapeutics in cancer. Annu Rev Pharmacol
Toxicol. 51:25–43. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Slack FJ and Weidhaas JB: MicroRNA in
cancer prognosis. N Engl J Med. 359:2720–2722. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
O'Day E and Lal A: MicroRNAs and their
target gene networks in breast cancer. Breast Cancer Res.
12:2012010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang L, Huang Q, Zhang S, Zhang Q, Chang
J, Qiu X and Wang E: Hsa-miR-125a-3p and hsa-miR-125a-5p are
downregulated in non-small cell lung cancer and have inverse
effects on invasion and migration of lung cancer cells. BMC Cancer.
10:3182010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim
JH, Kim JW and Kim S: MicroRNA expression profiles in serous
ovarian carcinoma. Clin Cancer Res. 14:2690–2695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tong Z, Liu N, Lin L, Guo X, Yang D and
Zhang Q: miR-125a-5p inhibits cell proliferation and induces
apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1.
Biomed Pharmacother. 75:129–136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang W, Qian P, Zhang X, Zhang M, Wang H,
Wu M, Kong X, Tan S, Ding K, Perry JK, et al: Autocrine/paracrine
human growth hormone-stimulated microRNA 96-182-183 cluster
promotes epithelial-mesenchymal transition and invasion in breast
cancer. J Biol Chem. 290:13812–13829. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hurst DR: Metastasis suppression by BRMS1
associated with SIN3 chromatin remodeling complexes. Cancer
Metastasis Rev. 31:641–651. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mei P, Bai J, Shi M, Liu Q, Li Z, Fan Y
and Zheng J: BRMS1 suppresses glioma progression by regulating
invasion, migration and adhesion of glioma cells. PLoS One.
9:e985442014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
You J, He X, Ding H and Zhang T: BRMS1
regulates apoptosis in non-small cell lung cancer cells. Cell
Biochem Biophys. 71:465–472. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Roberts MR, Hong CC, Edge SB, Yao S,
Bshara W, Higgins MJ, Freudenheim JL and Ambrosone CB: Case-only
analyses of the associations between polymorphisms in the
metastasis-modifying genes BRMS1 and SIPA1 and breast tumor
characteristics, lymph node metastasis, and survival. Breast Cancer
Res Treat. 139:873–885. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim HS, Lee KS, Bae HJ, Eun JW, Shen Q,
Park SJ, Shin WC, Yang HD, Park M, Park WS, et al: MicroRNA-31
functions as a tumor suppressor by regulating cell cycle and
epithelial-mesenchymal transition regulatory proteins in liver
cancer. Oncotarget. 6:8089–8102. 2015.PubMed/NCBI
|
|
39
|
Xu J, Wang T, Cao Z, Huang H, Li J, Liu W,
Liu S, You L, Zhou L, Zhang T and Zhao Y: MiR-497 downregulation
contributes to the malignancy of pancreatic cancer and associates
with a poor prognosis. Oncotarget. 5:6983–6993. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang Q, Huang Z, Guo W, Ni S, Xiao X, Wang
L, Huang D, Tan C, Xu Q, Zha R, et al: microRNA-202-3p inhibits
cell proliferation by targeting ADP-ribosylation factor-like 5A in
human colorectal carcinoma. Clin Cancer Res. 20:1146–1157. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Orom UA, Nielsen FC and Lund AH:
MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and
enhances their translation. Mol Cell. 30:460–471. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tsai NP, Lin YL and Wei LN: MicroRNA
mir-346 targets the 5′-untranslated region of receptor-interacting
protein 140 (RIP140) mRNA and up-regulates its protein expression.
Biochem J. 424:411–418. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vasudevan S, Tong Y and Steitz JA:
Switching from repression to activation: microRNAs can up-regulate
translation. Science. 318:1931–1934. 2007. View Article : Google Scholar : PubMed/NCBI
|