Introduction
The incidence and mortality rates of lung cancer are
the highest among all types of malignant neoplasm in China and
worldwide (1–3). Lung cancer can be divided into two
histological categories: Non-small cell lung cancer (NSCLC) and
small cell lung cancer (SCLC). NSCLC accounts for ~80% of lung
cancer cases, including lung adenocarcinoma (LUAD), lung squamous
cell carcinoma (LUSC) and lung large cell carcinoma (LULC)
(4,5).
Although the incidence of LUSC can be prevented to some extent by
regulating tobacco smoking (6), the
mechanism of tumorigenesis and progression of LUSC remains poorly
characterized. There has been great progress in molecular targeted
therapies for LUSC (7–9). However, resistance to targeted drugs
remains high, and the proportion of responsive patients is limited.
Therefore, the development of novel diagnostic and therapeutic
tools for LUSC is of great importance.
microRNA (miRNA) is a class of endogenous non-coding
RNA molecules, which are ~22 nucleotides in length and regulate
protein translation (10,11). Previous studies have demonstrated that
miRNAs silence genes by inhibiting the synthesis of associated
proteins or degrading the mRNA of target genes, and therefore serve
an important role in the regulation of growth, proliferation,
differentiation, apoptosis and development of cancer (12–14).
Numerous studies have reported the role of miRNA in the progression
of LUSC and its value in the diagnosis, treatment and prognosis of
this disorder (15–18).
miR-452-5p (previously named miR-452) is obtained by
modification of the 5′ end of pre-miR-452 (19,20).
Previous studies have reported that miR-452-5p expression in
different types of cancer is distinct, where its expression is
upregulated in urinary tract epithelial tumors, esophageal cancer
and liver cancer (21–23) but downregulated in breast cancer,
prostate cancer and glioma (24–26). Zhang
et al (27) and He et
al (28,29) reported low expression of miR-452-5p in
NSCLC cells and tissues, respectively. However, these studies were
limited by the quality of the LUSC samples used. Therefore, the
expression level and clinical significance of miR-452-5p in LUSC
remain unclear. The Cancer Genome Atlas (TCGA) and the Gene
Expression Omnibus (GEO) aim to provide a comprehensive
understanding of cancer at the molecular level using genomic
analyses, including large-scale genome sequences and microarrays
for improving the diagnosis, treatment and prevention of cancer. In
the present study, the expression level of miR-452-5p in LUSC was
confirmed using the GEO and TCGA databases for the purpose of
reducing the errors caused by sample size and study population.
Gene ontology (GO) and pathway enrichment analyses of miR-452-5p
target genes were also performed for clarification of the molecular
mechanism of miR-452-5p in LUSC.
Materials and methods
Extraction of data on miR-452-5p
expression in LUSC from TCGA and GEO
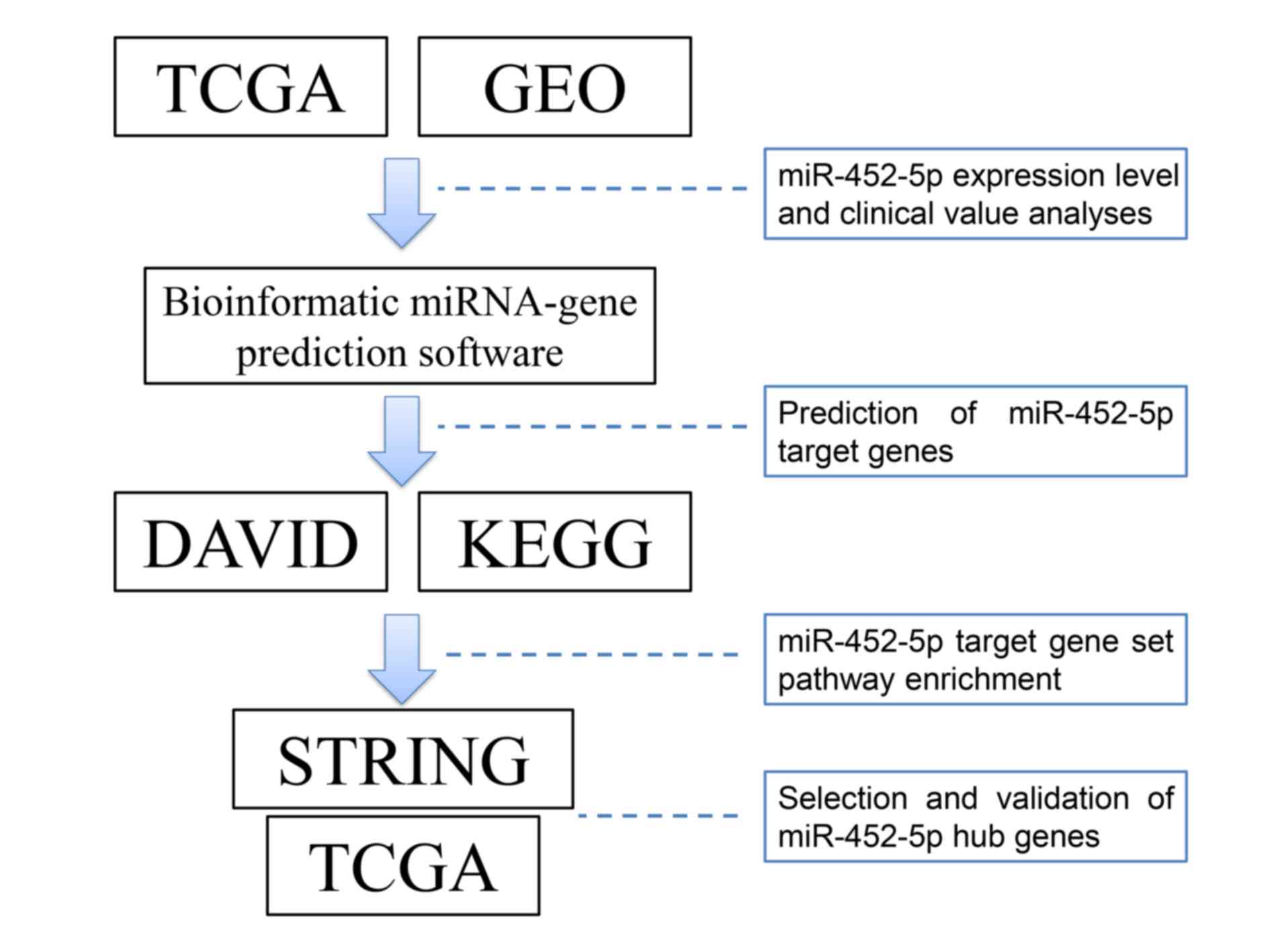
As illustrated in Fig.
1, information regarding miR-452-5p expression and clinical
characteristics of patients with LUSC was extracted from TCGA
(https://tcga-data.nci.nih.gov/docs/publications/tcga/)
and GEO (https://www.ncbi.nlm.nih.gov/geo/) for this
retrospective study. The following strategy was used to search in
GEO datasets: ‘Lung’ OR ‘pulmonary’ OR ‘respiratory’ OR ‘bronchi’
OR ‘alveoli’ AND ‘cancer’ OR ‘carcinoma’ OR ‘tumor’ OR ‘neoplas*’
OR ‘malignan*’. The resulting eligible records, including miRNA
microarray and RNA-seq datasets, were reviewed. The inclusion
criteria were as follows: i) Diagnosis of patient with lung
squamous cell carcinoma; ii) detection of miR-452-5p expression
level in tissue or blood samples and iii) availability of original
expression profiling data of miR-452-5p in both cancerous and
non-cancerous specimens. The exclusion criteria were as follows: i)
Datasets from research on cell lines or animals; ii) cancerous or
non-cancerous groups with small sample sizes (n<10), and iii)
poor-quality profiling expression data (0, 0.1 or 1) that accounts
for >30% of the total expression data. Independent investigators
(Xiaoning Gan and Tingqing Gan) reviewed the datasets that met the
criteria and extracted the appropriate datasets. Discrepancies
between the decisions of the two investigators were resolved by
discussion among all authors. Next, the data were summarized and
analyzed using Microsoft Office 2007 software package (Excel and
Office programmes), SPSS (version 22.0; IBM Corp., Armonk, NY, USA)
and R (version 3.3.0; https://www.r-project.org/). The primary expression
data of miR-452-5p were log2-transformed for further study.
Prediction of miR-452-5p target
genes
The prediction was performed using online
bioinformatic software, including EMBL-EBI (https://www.ebi.ac.uk/), Targetminer (https://www.isical.ac.in/~bioinfo_miu/targetminer20.htm),
DIANA-microT (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php),
miRWalk and databases associated with miRWalk (TargetScan, miRanda,
Pictar2, RNAhybrid, miRDB, RNA22-HAS, TargetMiner, EMBL-EBI,
DIANA-microT, mirbridge, miRMap, miRNAMap and PITA). The target
genes of miR-452-5p were searched in the PubMed (https://www.ncbi.nlm.nih.gov/pubmed) and EMBASE
(https://www.embase.com) databases using the
terms: ‘Cancer’, ‘tumor’, ‘carcinoma’, ‘neoplasm’, ‘malignant’,
‘malignancy’ and ‘miR-452’. Potential miR-452-5p target genes that
were positive in all seven software programs of the 14
aforementioned prediction software or have been previously
experimentally confirmed were used for further analyses.
Bioinformatic analysis of miR-452-5p
target genes
Potential miR-452-5p target genes were subjected to
GO and pathway analyses using the Database for Annotation,
Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/) and EnrichmentMap (a
Cytoscape plugin) (30). The Kyoto
Encyclopedia of Genes and Genomes database (KEGG; http://www.kegg.jp/) was used to identify the
signaling pathways associated with the target genes. The Search
Tool for the Retrieval of Interacting Genes/Proteins (STRING;
http://string-db.org/) database was utilized for
the selection of hub genes that were most likely involved in the
strategic pathway associated with LUSC. Hub genes were identified
by the combined score summarized in the protein-protein interaction
(PPI) network of STRING with Cytoscape (31,32).
Statistical analyses
Statistical analyses were performed using SPSS
(version 22.0; IBM Corp.), StataSE (version 12.0; StataCorp LP,
College Station, TX, USA), GraphPad Prism (version 5.0; GraphPad
Software Inc., La Jolla, CA, USA), R version 3.3.0 and Microsoft
Office 2007 software packages. Student's t-test (Paired or unpaired
t-test were utilized according to the data type analyzed) or the
Mann-Whitney test was used to examine the difference between the 2
groups. One-way analysis of variance followed by
Student-Newman-Keuls (SNK) post-hoc test was employed to compare
between ≥3 groups. Spearman's rank correlation coefficient analysis
was applied to analyze the correlation between miR-452-5p
expression and clinicopathological parameters or hub genes.
Receiver operating characteristic (ROC) curve analysis was used to
evaluate the diagnostic value of miR-452-5p in patients with LUSC.
The effect of miR-452-5p expression and clinical risk factors on
the survival of patients with LUSC was statistically analyzed using
the Cox proportional hazards regression model. A meta-analysis was
performed using StataSE (version 12.0). A forest plot of standard
mean deviation (SMD) was analyzed to confirm the expression level
of miR-452-5p in LUSC. If the standard mean deviation (SMD) was
<0 and the 95% CI was <0, the expression of miR-452-5p was
considered to be lower in cancerous specimens compared with
non-cancerous specimens. By contrast, if the overall SMD was >0
and the 95% CI was >0, the expression of miR-452-5p was
considered to be higher in cancerous specimens compared with
non-cancerous specimens. Publication bias was assessed using Begg's
funnel plot and Egger's test. P<0.05 was considered to indicate
a statistically significant difference.
Results
Association between miR-452-5p
expression and clinicopathological features in LUSC
The data on 387 cases of LUSC (342 cancerous and 45
non-cancerous adjacent lung tissues) patients with differential
miR-452-5p expression were extracted from the TCGA database. The
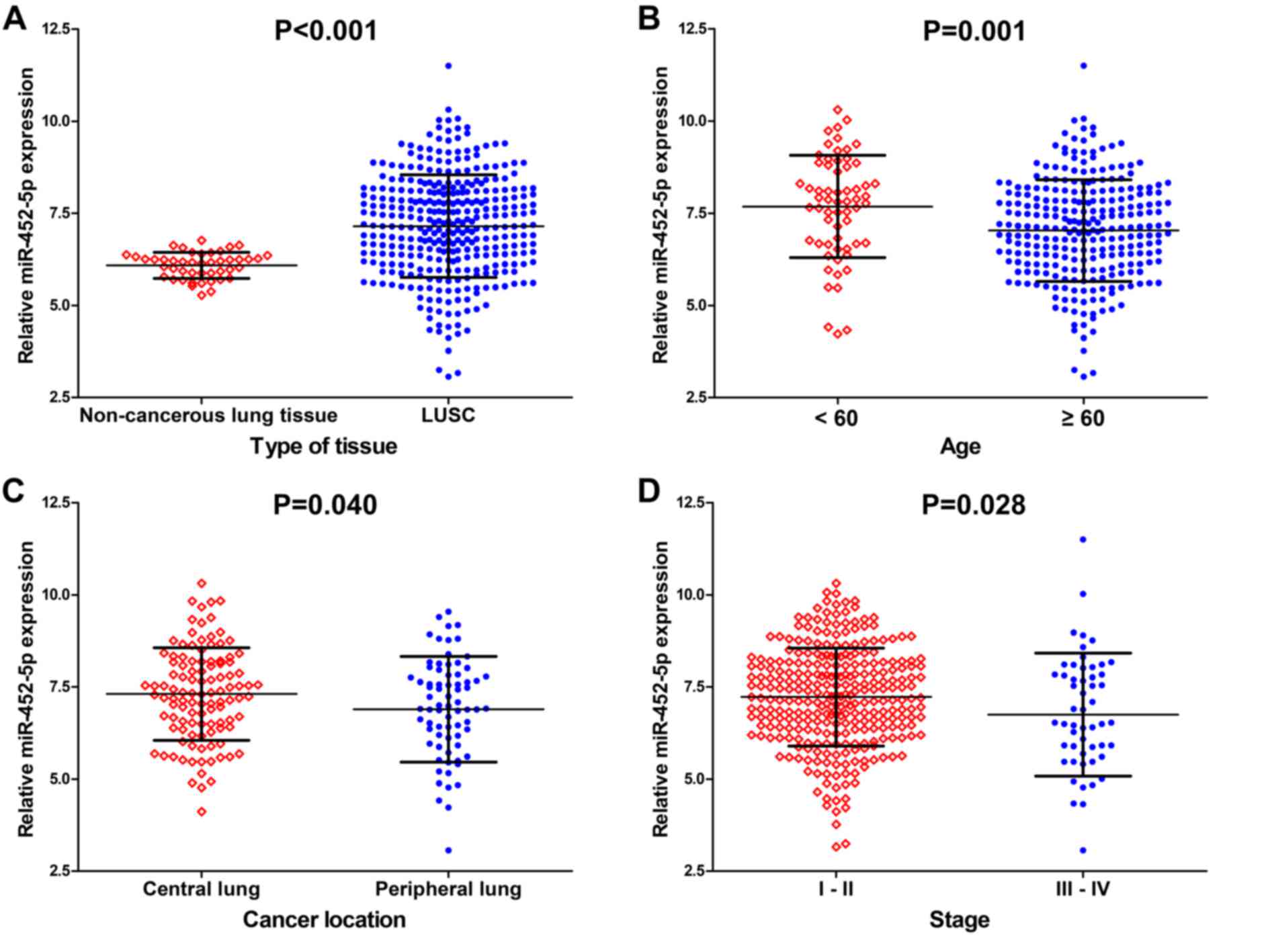
results indicated that the expression of miR-452-5p in LUSC tissues
was higher compared with adjacent lung tissues (7.1525±1.39063 vs.
6.0885±0.35298; P<0.001; Fig. 2A;
Table I).
 | Table I.Association between the expression of
miR-452-5p and clinicopathological parameters in LUSC as analyzed
using data from The Cancer Genome Atlas. |
Table I.
Association between the expression of
miR-452-5p and clinicopathological parameters in LUSC as analyzed
using data from The Cancer Genome Atlas.
|
|
| miR-452-5p
expression (log2) |
|---|
|
|
|
|
|---|
| Clinicopathological
features | n | Mean ± SD | t-test | P-value |
|---|
| Tissue |
|
|
|
|
|
Adjacent non-cancerous lung
tissue | 45 | 6.0885±0.35298 |
Z=−6.053a | <0.001 |
|
LUSC | 342 | 7.1525±1.39063 |
|
|
| Age (years) |
|
|
|
|
|
<60 | 62 | 7.6847±1.39161 | 3.359 | 0.001 |
|
≥60 | 274 | 7.0333±1.37623 |
|
|
| Sex |
|
|
|
|
|
Female | 85 | 7.0867±1.30046 | −0.503 | 0.616 |
|
Male | 257 | 7.1742±1.42095 |
|
|
| Ethnicity |
|
|
|
|
|
Caucasian descent | 252 | 7.1228±1.42366 |
F=1.985b | 0.139 |
| African
descent | 24 | 7.4428±1.32204 |
|
|
| Asian
descent | 6 | 6.1575±1.78242 |
|
|
| Recurrence |
|
|
|
|
| No | 139 | 7.2663±1.29109 | −0.315 | 0.753 |
|
Yes | 30 | 7.3517±1.57974 |
|
|
| Cancer
location |
|
|
|
|
| Central
lung | 109 | 7.3056±1.25173 | 2.069 | 0.040 |
|
Peripheral lung | 74 | 6.8924±1.42939 |
|
|
| Smoke |
|
|
|
|
|
≤20 | 29 | 7.2479±1.77796 | 0.379 | 0.705 |
|
>20 | 258 | 7.1428±1.37157 |
|
|
| Stage |
|
|
|
|
|
I–II | 285 | 7.2259±1.32747 |
Z=−2.191a | 0.028 |
|
III–IV | 54 | 6.7484±1.67113 |
|
|
| Tumor grade |
|
|
|
|
|
T1-2 | 272 | 7.1847±1.36195 | 0.844 | 0.399 |
|
T3-4 | 70 | 7.0273±1.50065 |
|
|
| Nodal
metastasis |
|
|
|
|
| No | 219 | 7.2583±1.26109 |
Z=−1.901a | 0.057 |
|
Yes | 123 | 6.9642±1.58355 |
|
|
| Metastasis |
|
|
|
|
| No | 261 | 7.1327±1.35815 | −0.471 | 0.638 |
|
Yes | 81 | 7.2161±1.49758 |
|
|
| Tumor status |
|
|
|
|
|
Tumor-free | 216 | 9.2886±1.28058 | −0.682 | 0.496 |
| With
tumor | 57 | 9.4231±1.48230 |
|
|
The associations between the levels of miR-452-5p
expression and age, cancer location, and TNM stage were
statistically significant. miR-452-5p expression in patients <60
years was significantly increased compared with patients ≥60 years
(P=0.001; Fig. 2B). The expression of
miR-452-5p in patients with a tumor located in the central lung was
significantly increased compared with patients with a tumor located
in the peripheral lung (P=0.040; Fig.
2C). Additionally, the level of miR-452-5p expression in
patients with TNM stages I–II LUSC was significantly increased
compared with patients with patients with stages III–IV LUSC
(P=0.028; Fig. 2D). Spearman's rank
correlation coefficient analysis demonstrated that the expression
of miR-452-5p was negatively correlated with age (r=−0.187;
P=0.001) and TNM stage (r=−0.119; P=0.028) but was not
significantly correlated with other clinical parameters.
Diagnostic value of miR-452-5p
expression in LUSC
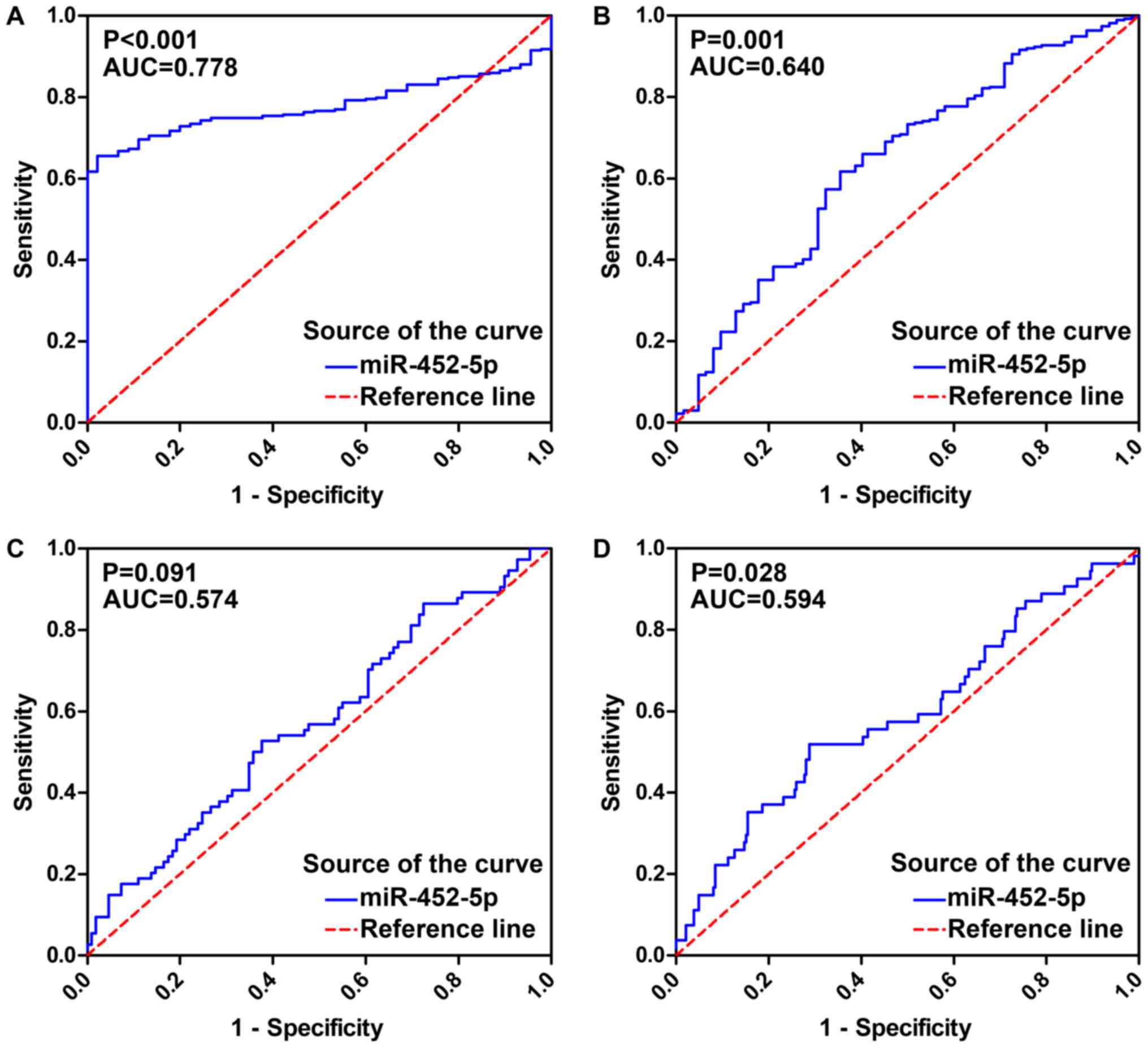
An ROC was generated to verify the predictive
performance of miR-452-5p expression levels and various clinical
parameters in LUSC (Fig. 3). The AUC
of miR-452-5p expression for the diagnosis of LUSC was 0.778 (95%
CI, 0.734–0.821; P<0.001) with a sensitivity of 65.5%, a
specificity of 97.8% and a diagnostic threshold value of 6.632. The
AUC of miR-452-5p expression for age was 0.640 (95% CI,
0.561–0.718; P=0.001). Furthermore, in LUSC patients with different
tumor location and TNM stage, the AUC values were 0.574 (95% CI,
0.489–0.658; P=0.091) and 0.594 (95% CI, 0.507–0.681; P=0.028),
respectively.
Prognostic value of miR-452-5p in
LUSC
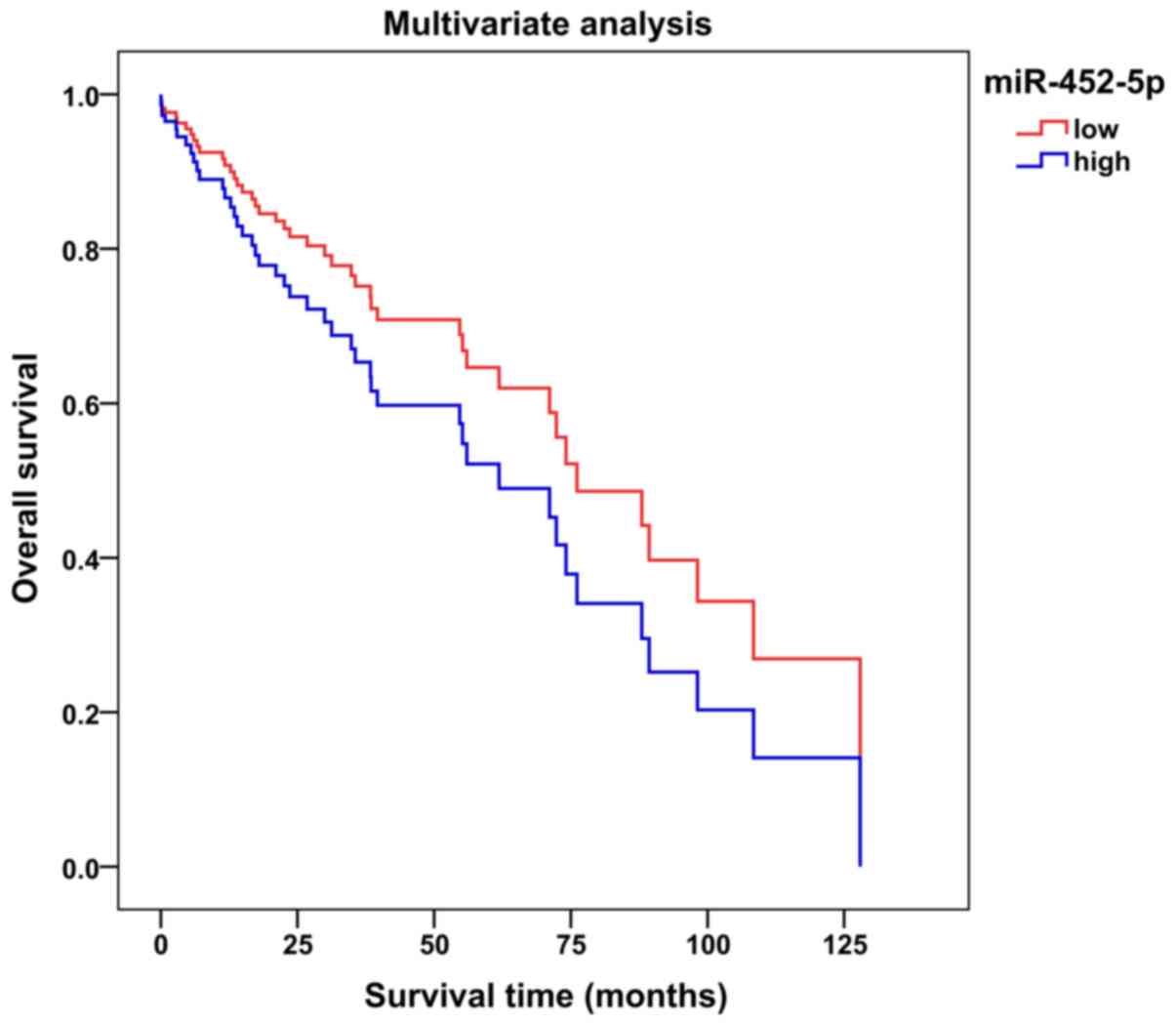
An average value of 7.1525 was selected as the
miR-452-5p expression level threshold to evaluate the association
between miR-452-5p expression levels and the survival of patients
with LUSC. The multivariate survival curve suggested that patients
with LUSC and a low miR-452-5p expression may have longer overall
survival (time from randomization until mortality from any cause)
compared with patients with LUSC and a high miR-452-5p expression
(Fig. 4; Table II). It was not possible to determine
whether miR-452-5p could be a prognostic factor in patients with
LUSC [univariate hazard ratio (HR), 0.981; 95% CI, 0.648–1.487;
P=0.928; multivariate HR, 1.492; 95% CI, 0.791–2.814; P=0.216].
 | Table II.Multivariate analysis of
clinicopathological parameters for overall survival in lung
squamous cell carcinoma. |
Table II.
Multivariate analysis of
clinicopathological parameters for overall survival in lung
squamous cell carcinoma.
| A, Univariate
analysis |
|---|
|
|---|
| Clinicopathological
features | HR | 95% CI | P-value |
|---|
| Age | 1.163 | 0.734–1.844 | 0.521 |
| >3
vs. ≤3 |
|
|
|
| Tumor grade | 1.489 | 1.016–2.182 | 0.041 |
| T3-4
vs. T1-2 |
|
|
|
| Nodal
metastasis |
|
|
|
| Yes vs.
no | 1.222 | 0.886–1.686 | 0.221 |
| Metastasis |
|
|
|
| Yes vs.
no | 1.343 | 0.836–2.156 | 0.222 |
| TNM stage |
|
|
|
| III–IV
vs. I–II | 1.441 | 1.001–2.075 | 0.049 |
| Cancer
location |
|
|
|
|
Peripheral lung vs. central
lung | 1.242 | 0.768–2.010 | 0.377 |
| miR-452-5p
expression |
|
|
|
| High
vs. low | 0.981 | 0.648–1.487 | 0.928 |
|
| B, Multivariate
analysis |
|
|
Clinicopathological features | HR | 95% CI | P-value |
|
| Age | 1.596 | 0.535–4.760 | 0.401 |
| >3
vs. ≤3 |
|
|
|
| Tumor grade | 0.999 | 0.416–2.396 | 0.998 |
| T3-4
vs. T1-2 |
|
|
|
| Nodal
metastasis |
|
|
|
| Yes vs.
no | 1.307 | 0.611–2.793 | 0.490 |
| Metastasis |
|
|
|
| Yes vs.
no | 1.358 | 0.301–6.126 | 0.691 |
| TNM stage |
|
|
|
| III–IV
vs. I–II | 1.358 | 0.566–3.259 | 0.494 |
| Cancer
location |
|
|
|
|
Peripheral lung vs. central
lung | 1.943 | 0.973–3.884 | 0.060 |
| miR-452-5p
expression |
|
|
|
| High
vs. low | 1.492 | 0.791–2.814 | 0.216 |
Verification of miR-452-5p expression
in LUSC with meta-analysis
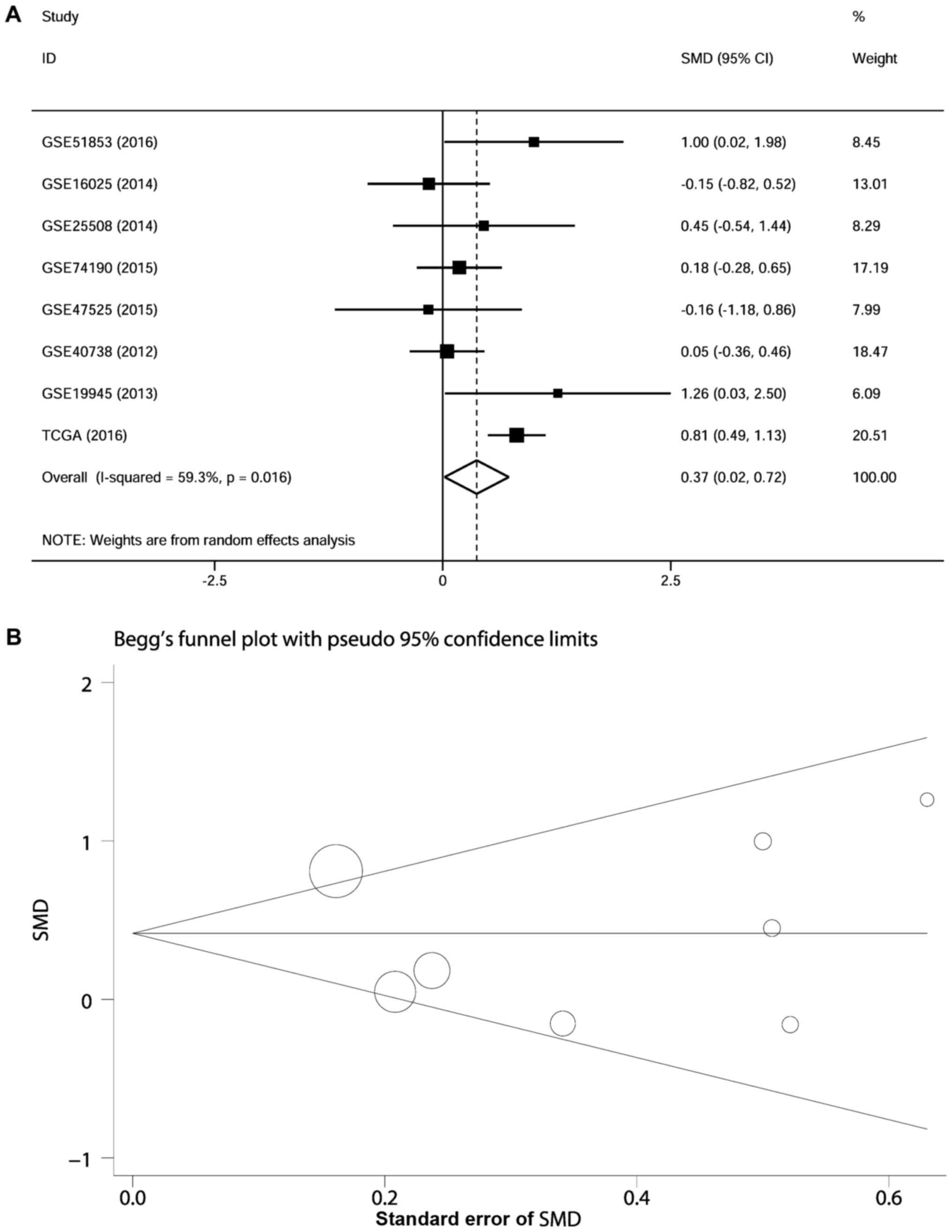
A total of 720 cancerous and non-cancerous samples
from 7 eligible GEO datasets (including 333 cases from 7 eligible
GEO datasets and 387 cases from TCGA) were screened in the
meta-analysis (data not shown). The miR-452-5p expression levels in
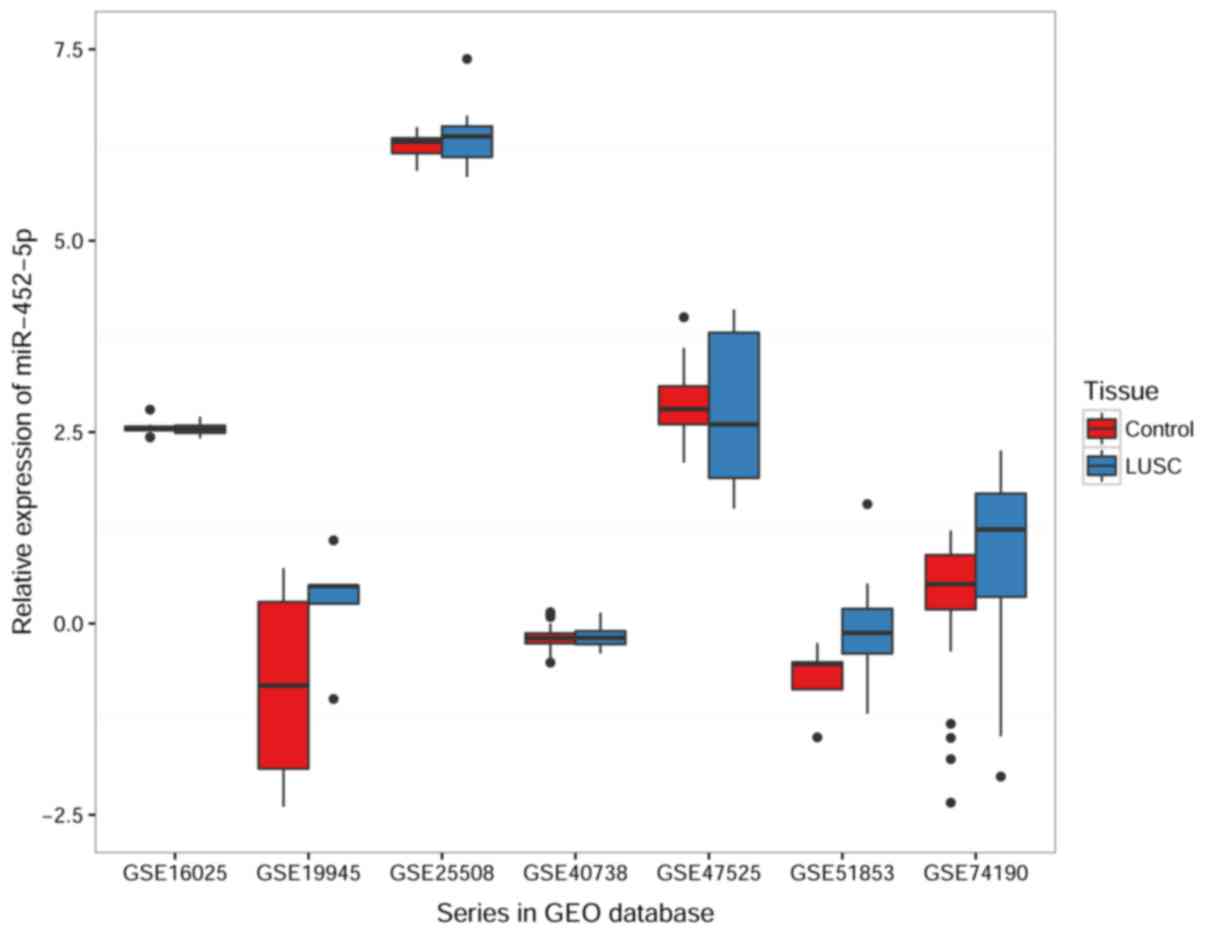
LUSC of each GEO dataset are illustrated in Fig. 5 and shown in Table III. In the meta-analysis, the
random-effects model was used to calculate the pooled SMD and 95%
CI due to the high heterogeneity of the data (I2=59.3%;
P=0.016). These results indicated that the expression of miR-452-5p
in LUSC was significantly increased compared with non-cancerous
specimens (SMD, 0.372; 95% CI, 0.020–0.724; z=2.07; P=0.038)
(Fig. 6A). The points, which
represent individual studies, were symmetrically arranged in the
Begg's funnel plot (P=0.711) and the P-values calculate from the
Egger's test were >0.05 (P= 0.810), which indicates that there
was no publication bias in the studies (Fig. 6B).
 | Table III.Relative expression of miR-452-5p in
LUSC analyzed using Gene Expression Omnibus datasets. |
Table III.
Relative expression of miR-452-5p in
LUSC analyzed using Gene Expression Omnibus datasets.
|
|
|
| miR-452-5p
expression (log2) |
|---|
|
|
|
|
|
|---|
| Dataset | Tissue | n | Mean ± SD | t-test | P-value |
|---|
| GSE40738 | Non-cancer | 72 |
−0.1856±0.12812 | −0.226 | 0.822 |
|
| LUSC | 34 |
−0.1796±0.12344 |
|
|
| GSE19945 | Non-cancer | 8 |
−1.3640±1.51488 | −2.213 | 0.049b |
|
| LUSC | 5 | 0.2678±0.76523 |
|
|
| GSE16025 | Non-cancer | 10 | 2.5530±0.10050 | 0.447 | 0.656 |
|
| LUSC | 61 | 2.5420±0.06697 |
|
|
| GSE25508 | Non-cancer | 8 | 6.2369±1.86325 | −1.085a | 0.314 |
|
| LUSC | 8 | 6.3974±0.46815 |
|
|
| GSE74190 | Non-cancer | 44 |
−0.4829±1.84267 | −0.769 | 0.445 |
|
| LUSC | 30 |
−0.1329±2.03639 |
|
|
| GSE47525 | Non-cancer | 14 | 2.8929±0.51361 | 0.213 | 0.840 |
|
| LUSC | 5 | 2.7800±1.14324 |
|
|
| GSE51853 | Non-cancer | 5 |
−0.7281±0.47630 | −2.062 | 0.047b |
|
| LUSC | 29 |
−0.1516±0.59057 |
|
|
Potential target genes of
miR-452-5p
A total of 17,094 miR-452-5p candidate target genes
were screened using 14 prediction programs. A total of 249
miR-452-5p target genes were searched and subjected to GO and
pathway analyses. This included 248 candidates that were verified
as target genes by ≥7 programs and 1 target gene that was validated
by experiments.
GO and KEGG pathway analyses
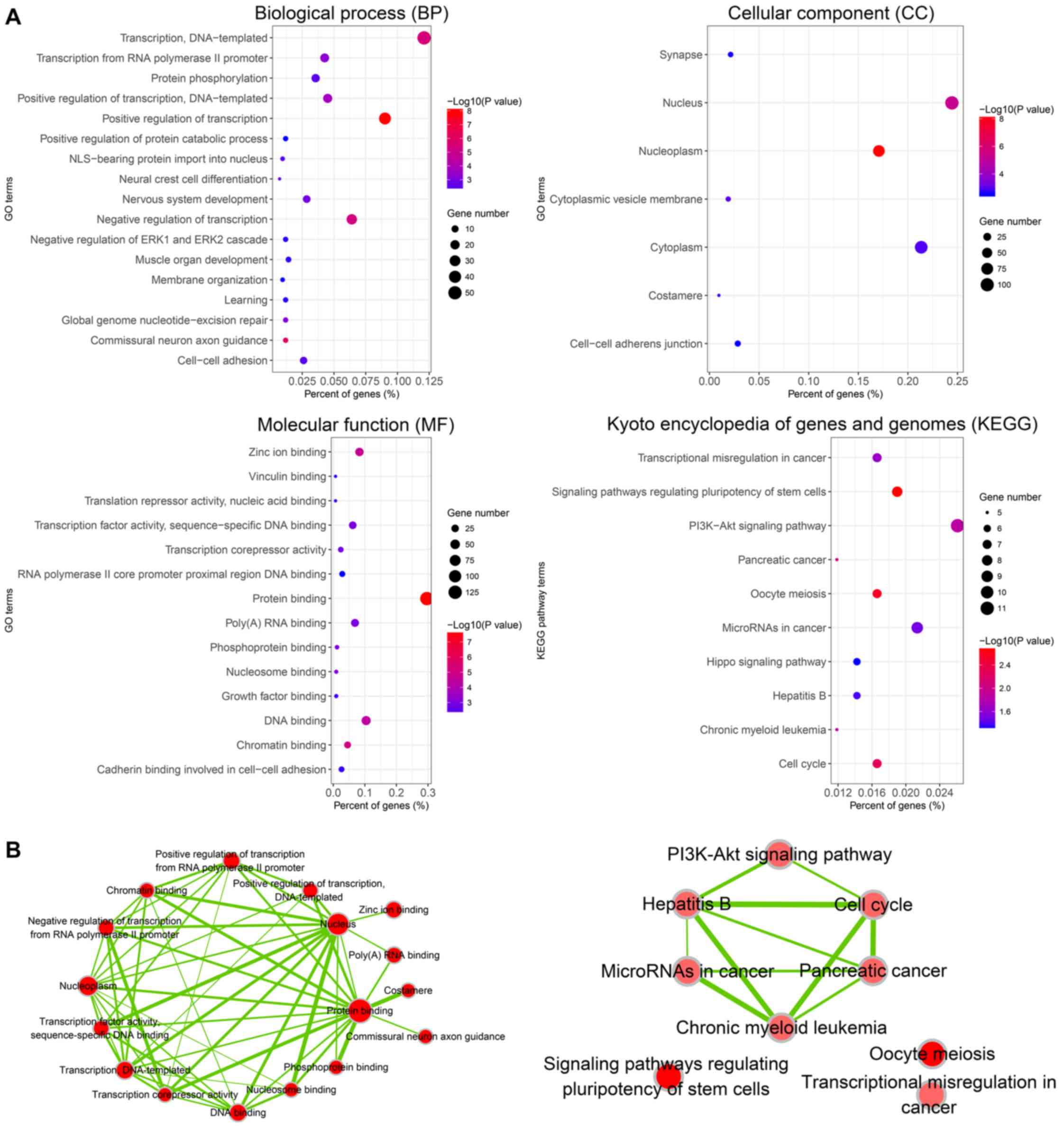
The DAVID database was used to perform GO analysis
of the 249 target genes of miR-452-5p (Fig. 7A). The results indicated that, at the
level of biological process (BP), target gene sets of miR-452-5p
were closely associated with ‘regulation of transcription’,
‘commissural neuron axon guidance’ and ‘transcription’ (P<0.01).
In terms of cellular components (CC), the target genes were mainly
involved in the assembly of cellular structures, including
‘nucleoplasm’, ‘nucleus’ and ‘cytoplasmic vesicle membrane’
(P<0.01). Regarding molecular function (MF), the genes were
mainly enriched in protein binding, chromatin binding and zinc ion
binding (P<0.01). The results from the KEGG pathway analysis
indicated that the predicted miR-452-5p target gene sets were
significantly enriched in signaling pathways that are involved in
the regulation of pluripotency of stem cells, oocyte meiosis and
cell cycle (P<0.05).
The visualization function of Cytoscape was used to
graphically display the results of GO (P<0.01; Q<0.1; overlap
coefficient >0.5) and KEGG (P<0.05; Q<0.5; overlap
coefficient >0.5) enrichment annotation analyses, allowing a
more intuitive presentation of the associations between the
pathways (Fig. 7B).
PPI network analysis for the selection
of hub genes
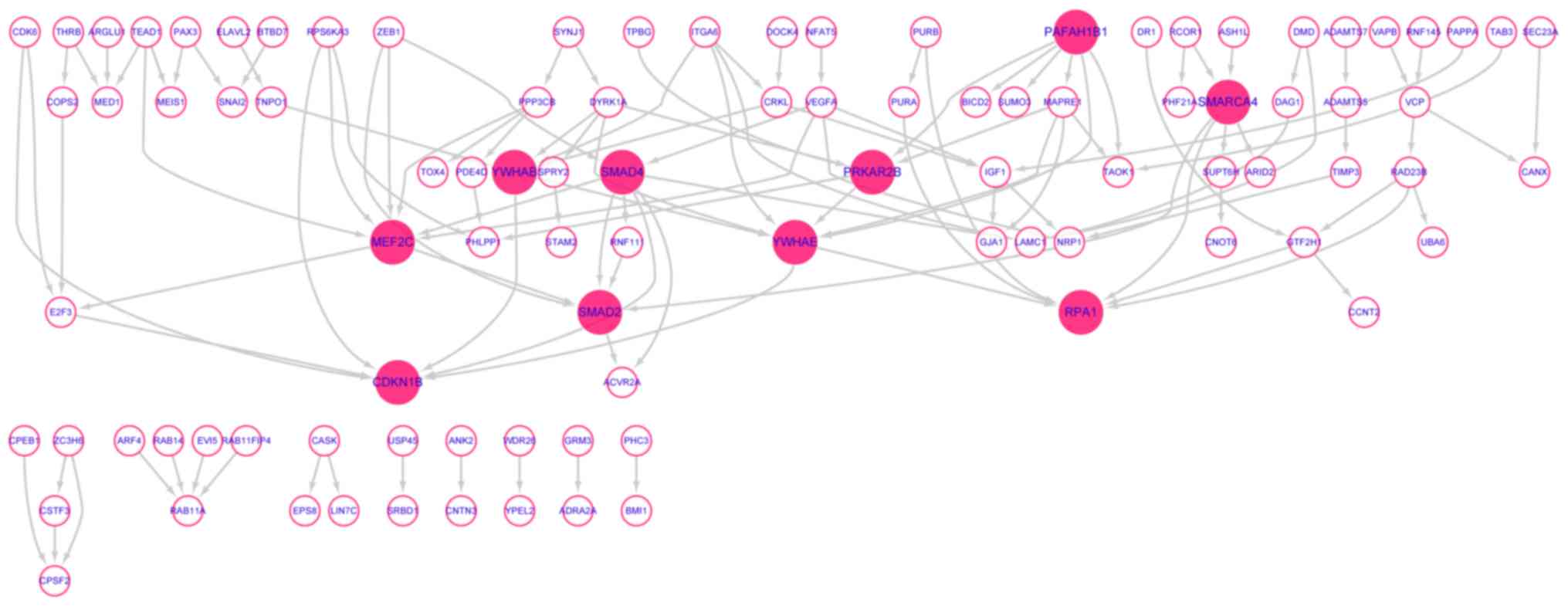
The network consisted of 249 nodes and 118 edges.
Among the 249 miR-452-5p target genes, 95 target genes were
connected based on experiment, co-expression, neighborhood or other
evidence summarized by STRING. The disconnected target genes
(n=154) were removed from the network (Fig. 8). Finally, 10 hub genes (nodes
connected with more than 6 edges) were identified from the 95
target genes. These hub genes were SMAD family member 4 (SMAD4),
myocyte enhancer factor 2C (MEF2C), tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein ε
(YWHAE), SMAD family member 2 (SMAD2), SWI/SNF related, matrix
associated, actin-dependent regulator of chromatin, subfamily a,
member 4 (SMARCA4), tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein β (YWHAB), platelet activating
factor acetylhydrolase 1b regulatory subunit 1 (PAFAH1B1), protein
kinase cAMP-dependent type II regulatory subunit β (PRKAR2B),
cyclin dependent kinase inhibitor 1B (CDKN1B) and replication
protein A1 (RPA1). Among these 10 hub genes, 5 hub genes, including
SMAD4, SMAD2, CDKN1B, YWHAE and YWHAB, were significantly enriched
in ‘cell cycle’.
Association between the expression of
miR-452-5p and the 5 hub genes that are involved in cell cycle
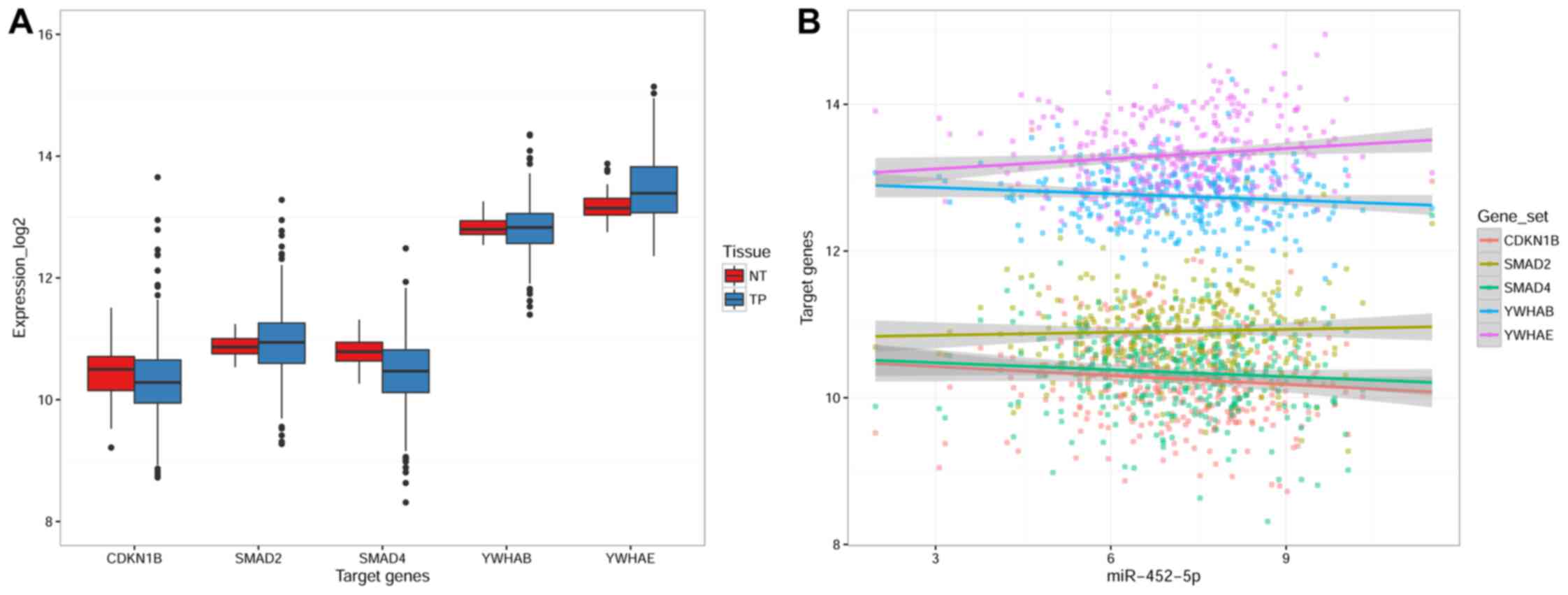
The expression levels of 5 hub genes in LUSC were
demonstrated with the TCGA dataset, including SMAD4, SMAD2, CDKN1B,
YWHAE and YWHAB. The results indicated that the expression of
CDKN1B and SMAD4 was significantly downregulated in LUSC tissues
compared with non-cancerous lung tissues (P=0.023 and P<0.001,
respectively), while YWHAE expression was significantly upregulated
in LUSC compared with control (P<0.001). There was no
significant difference in the expression of SMAD2 or YWHAB in LUSC
tissues compared with non-cancerous lung tissue (Fig. 9A; Table
IV). Furthermore, Spearman's rank correlation coefficient
analysis demonstrated that miR-452-5p expression was negatively
correlated with CDKN1B expression in LUSC (r=−0.163; P=0.003).
However, miR-452-5p expression was not significantly correlated
with the expression of SMAD4, SMAD2, YWHAE or YWHAB (Fig. 9B). In addition, in non-cancerous lung
tissue, miR-452-5p was not correlated with CDKN1B expression
(r=−0.013; P=0.937). These results suggested that miR-452-5p may
target CDKN1B in the carcinogenesis of LUSC.
 | Table IV.Relative expression of 5 hub target
genes of miR-452-5p in LUSC analyzed using data from The Cancer
Genome Atlas. |
Table IV.
Relative expression of 5 hub target
genes of miR-452-5p in LUSC analyzed using data from The Cancer
Genome Atlas.
|
|
| Hub gene relevant
expression (log2) |
|---|
|
|
|
|
|---|
| Groups | n | Mean ± SD | z | P-value |
|---|
| CDKN1B |
|
|
|
|
|
Non-cancer | 52 |
10.4490±0.47049 | −2.277 | 0.023a |
|
LUSC | 511 |
10.3141±0.60619 |
|
|
| SMAD2 |
|
|
|
|
|
Non-cancer | 52 |
10.8716±0.18524 | −1.076 | 0.282 |
|
LUSC | 511 |
10.9439±0.54278 |
|
|
| SMAD4 |
|
|
|
|
|
Non-cancer | 52 |
10.8014±0.22412 | −5.275 |
<0.001a |
|
LUSC | 511 |
10.4572±0.55275 |
|
|
| YWHAB |
|
|
|
|
|
Non-cancer | 52 |
12.8296±0.16600 | −0.259 | 0.796 |
|
LUSC | 511 |
12.8083±0.40408 |
|
|
| YWHAE |
|
|
|
|
|
Non-cancer | 52 |
13.1903±0.25322 | −3.541 |
<0.001a |
|
LUSC | 511 |
13.4415±0.52233 |
|
|
Discussion
The present study indicated that miR-452-5p
expression in LUSC was significantly upregulated compared with
non-cancerous lung tissues, and the diagnostic value of miR-452-5p
expression for LUSC was also confirmed. Correlation analysis
indicated that the expression of miR-452-5p correlated with
clinical parameters, including age and TNM stage. Additionally, the
results indicate that patients with LUSC and high expression of
miR-452-5p were associated with early tumor stage but shorter
survival time. There are two potential reasons that may lead to
these apparent conflicting results. Firstly, patients with
relatively higher expression of miR-452-5p were more likely to
succumb to disease at the early stage, while patients with relative
lower expression of miR-452-5p were more likely to survive until
late stages of the disease. Secondly, a number of patients with
high expression of miR-452-5p were unable to accept or refused the
investigation due to severe symptoms caused by the disease. These
findings indicate that miR-452-5p may serve an essential role in
the carcinogenesis and progression of LUSC. Furthermore, 5 hub
target genes of miR-452-5p that are known to be involved in the
cell cycle were predicted and screened, which may guide future
studies in elucidating the mechanism of LUSC. The results also
demonstrate that among these 5 hub genes, the expression of CDKN1B
was negatively correlated with miR-452-5p.
miRNAs mainly perform their biological functions by
complete or incomplete complementary binding with the
3′-untranslated region (UTR) of mRNAs (33–37).
miR-452-5p is located at Xq28 in humans, and it can target multiple
genes and thus serves a potentially important role in the
biological process of carcinogenesis via a variety of mechanisms.
Hu et al (24) reported that
miR-452-5p expression was downregulated in breast cancer, which may
lead to resistance to adriamycin by targeting the insulin analogue,
growth factor receptor 1 (IGF-1R). The downregulation of miR-452-5p
may also lead to resistance to docetaxel by targeting the anaphase
promoting complex subunit 4 (APC4) (24). Liu et al (26) reported that the expression of
multifunctional stem cell regulatory factors, including SRY-box 2
(SOX2), was downregulated following upregulation of miR-452-5p
expression in glioma cell lines. Furthermore, Liu et al
(26) confirmed that the
downregulation of miR-452-5p in glioma cells and tissues was
correlated with promoter methylation. Zheng et al (23) reported high expression of miR-452-5p
in hepatocellular carcinomas, suggesting that miR-452-5p could
promote hepatocellular carcinoma by targeting CDKN1B (23). Zhang et al reported that
miR-452-5p inhibited the proliferation, invasion and migration of
NSCLC cells via the process of epithelial-mesenchymal transition
(27). He et al (28) suggested that miR-452-5p enhanced the
invasive capability of NSCLC cells by targeting BMI1
proto-oncogene, polycomb ring finger (28). However, the potential function and
mechanism of miR-452-5p expression in LUSC remain unclear.
To date, to the best of our knowledge, He et
al (29) is the only group to
have reported that the downregulated expression of miR-452-5p was
associated with clinicopathological features in patients with
NSCLC. Although the nature of the present research study was
similar to the study by He et al (29), the findings in our study are novel and
provide further insights. Firstly, the present study analyzed a
larger sample size using meta-analysis. The expression level of
miR-452-5p in LUSC was confirmed using a total of 720 samples from
TCGA and GEO, and whilst He et al (29) detected miR-452-5p expression in 76
LUSC samples, no association between high miR-452-5p expression and
LUSC was observed. Secondly, the clinical value of miR-452-5p as a
biomarker was considered in the present study, where it is
indicated that miR-452-5p may contribute to the diagnosis of LUSC.
Furthermore, miR-452-5p may be useful in predicting the progression
of LUSC due to the close association between miR-452-5p expression
and TNM stage. Finally, a systematic analysis of the potential
molecular mechanisms of miR-452-5p function in biological processes
that underlie LUSC was performed. A combination of 14 online
prediction tools were employed to screen target genes of
miR-452-5p. As a result, 249 miR-452-5p target genes were selected
for GO and pathway analyses, it was indicated that the target genes
primarily functioned by binding with protein, chromatin or other
biological molecules, which are involved in transcriptional
regulation.
Additionally, in order to analyze the key protein
involved in the carcinogenesis, protein-protein interaction (PPI)
analysis of 249 miR-452-5p target genes was applied. Consequently,
10 hub genes including SMAD4, SMAD2, CDKN1B, YWHAB, YWHAE, MEF2C,
SMARCA4, PAFAH1B1, PRKAR2B and RPA1 were identified for additional
study. Of these, the hub genes involved in cell cycle, including
SMAD4, SMAD2, CDKN1B, YWHAE and YWHAB, are of particular interest.
The cell cycle is a critical pathway that is closely associated
with the prognosis and therapy of numerous malignancies,
particularly LUSC (38,39). In addition, half of the 10 hub genes
were significantly enriched in ‘cell cycle’. Notably, CDKN1B has
been reported to function as a tumor suppressor gene in
hepatocellular carcinoma and it is targeted by miR-452-5p (23). Therefore, it was hypothesized if
CDKN1B, a hub gene which is involved in cell cycle, may have an
important role in molecular targeting therapy of LUSC. In the
present study, it was demonstrated that the expression of
miR-452-5p was negatively associated with the levels of CDKN1B in
LUSC and this association was not statistical significant in normal
lung tissues. These findings provide a direction for future
investigation of the molecular mechanism that underlies miR-452-5p
function in LUSC. Consistent with the results in the present study,
Zheng et al (23) reported
that miR-452-5p may directly target and suppress the expression of
CDKN1B in hepatocellular carcinoma (23). The study also indicated that
miR-452-5p may have a role in LUSC. Further experimental studies
that investigate the association between CDKN1B and miR-452-5p will
help to elucidate the mechanisms that underlie the carcinogenesis
of LUSC.
However, several limitations exist in the present
study. Firstly, heterogeneity may inevitably be present in the
meta-analysis of 720 samples, resulting from numerous factors,
including the detection method used, region, sex, age, stage and
the type of LUSC specimens (fresh frozen tissue, formalin-fixed
paraffin-embedded tissue or peripheral blood specimens). Therefore,
future evidence-based confirmation and subgroup analyses by
large-scale clinical trials are required to investigate the source
of the heterogeneity. Secondly, miRNAs regulate the function of
target genes during carcinogenesis and cancer progression through
different biological pathways, which form a complex regulatory
network of interactions involving biological molecules. Therefore,
the present study utilized bioinformatic methods to predict the
rudimentary function and mechanism of target genes. GO and pathway
analyses were conducted and a PPI network model of biological
molecules was constructed. However, in order to determine the
mechanisms of miR-452-5p involved in LUSC, further verification,
through in vitro and in vivo experiments as well as
clinical trials, is required to identify new targets for potential
treatments of LUSC patients.
In summary, relatively high levels of miR-452-5p
expression in LUSC compared with non-cancerous lung tissues were
determined by meta-analysis, which utilized experimental data
obtained from TCGA and GEO. In addition, high expression of
miR-452-5p was closely associated with the clinical parameters of
LUSC, including age, cancer location and TNM stage. Furthermore, it
is speculated that miR-452-5p serves a critical role in LUSC
carcinogenesis by targeting CDKN1B, which is involved in cell
cycle. Taken together, these findings provide a new direction for
the development of tools for early diagnosis and targeted treatment
of LUSC.
Acknowledgements
Not applicable.
Funding
The present study was supported by a fund from the
Guangxi Provincial Health Bureau Scientific Research Project (grant
nos. Z2013201 and Z2014055), the National Natural Science
Foundation of China (grant nos. NSFC81360327 and NSFC81560469), the
Natural Science Foundation of Guangxi, China (grant no.
2015GXNSFCA139009), the Scientific Research Project of the Guangxi
Education Agency (grant nos. KY2015 and LX062) and the Scientific
Research Project of the Basic Ability Promoting for Middle Age and
Youth Teachers of Guangxi Universities (grant no. KY2016YB077).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article
Authors' contributions
XG, TG, RH, JL, RT, HW, HZ, JM and HQ performed the
literature search, data extraction and statistical analysis, and
drafted the paper. GC and XH supervised the literature search, data
extraction and analysis, and reviewed the paper. All authors have
read and approved the manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of the First Affiliated Hospital of Guangxi Medical
University, with informed consent signed by all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McGuire S: World cancer report 2014.
Geneva, Switzerland: World health organization, international
agency for research on cancer, WHO Press, 2015. Adv Nutr.
7:418–419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hu M, Hu Y, He J and Li B: Prognostic
value of basic fibroblast growth factor (bFGF) in lung cancer: A
systematic review with meta-analysis. PLoS One. 11:e01473742016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schmidt B, Beyer J, Dietrich D, Bork I,
Liebenberg V and Fleischhacker M: Quantification of cell-free
mSHOX2 Plasma DNA for therapy monitoring in advanced stage
non-small cell (NSCLC) and small-cell lung cancer (SCLC) patients.
PLoS One. 10:e01181952015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takahashi A, Ishii G, Neri S, Yoshida T,
Hashimoto H, Suzuki S, Umemura S, Matsumoto S, Yoh K, Niho S, et
al: Podoplanin-expressing cancer-associated fibroblasts inhibit
small cell lung cancer growth. Oncotarget. 6:9531–9541. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meza R, Meernik C, Jeon J and Cote ML:
Lung cancer incidence trends by gender, race and histology in the
United States, 1973–2010. PLoS One. 10:e01213232015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun Y, Han Y, Wang X, Wang W, Wang X, Wen
M, Xia J, Xing H, Li X and Zhang Z: Correlation of EGFR Del 19 with
Fn14/JAK/STAT signaling molecules in non-small cell lung cancer.
Oncol Rep. 36:1030–1040. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee CK, Brown C, Gralla RJ, Hirsh V,
Thongprasert S, Tsai CM, Tan EH, Ho JC, Chu da T, Zaatar A, et al:
Impact of EGFR inhibitor in non-small cell lung cancer on
progression-free and overall survival: A meta-analysis. J Natl
Cancer Inst. 105:595–605. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Drilon A, Rekhtman N, Ladanyi M and Paik
P: Squamous-cell carcinomas of the lung: Emerging biology,
controversies, and the promise of targeted therapy. Lancet Oncol.
13:e418–e426. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu C, Li S, Chen T, Hu H, Ding C, Xu Z,
Chen J, Liu Z, Lei Z, Zhang HT, et al: miR-296-5p suppresses cell
viability by directly targeting PLK1 in non-small cell lung cancer.
Oncol Rep. 35:497–503. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu Y, Miao L, Ni R, Zhang H, Li L, Wang
X, Li X and Wang J: microRNA-520a-3p inhibits proliferation and
cancer stem cell phenotype by targeting HOXD8 in non-small cell
lung cancer. Oncol Rep. 36:3529–3535. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang D, Ma J, Ji X, Xu F and Wei Y:
miR-141 regulation of EIF4E expression affects docetaxel
chemoresistance of non-small cell lung cancer. Oncol Rep.
37:608–616. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou L, Di Q, Sun B, Wang X, Li M and Shi
J: MicroRNA-194 restrains the cell progression of non-small cell
lung cancer by targeting human nuclear distribution protein C.
Oncol Rep. 35:3435–3444. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen X, Wei L and Zhao S: miR-338 inhibits
the metastasis of lung cancer by targeting integrin β3. Oncol Rep.
36:1467–1474. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu X, Liu T, Fang O, Dong W, Zhang F,
Leach L, Hu X and Luo Z: MicroRNA-708-5p acts as a therapeutic
agent against metastatic lung cancer. Oncotarget. 7:2417–2432.
2016.PubMed/NCBI
|
|
16
|
Li D, Du X, Liu A and Li P: Suppression of
nucleosome-binding protein 1 by miR-326 impedes cell proliferation
and invasion in non-small cell lung cancer cells. Oncol Rep.
35:1117–1124. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Taylor MA, Wappett M, Delpuech O, Brown H
and Chresta CM: Enhanced MAPK signaling drives ETS1-mediated
induction of miR-29b leading to downregulation of TET1 and changes
in epigenetic modifications in a subset of lung SCC. Oncogene.
35:4345–4357. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Olbromski M, Grzegrzolka J,
Jankowska-Konsur A, Witkiewicz W, Podhorska-Okolow M and Dziegiel
P: MicroRNAs modulate the expression of the SOX18 transcript in
lung squamous cell carcinoma. Oncol Rep. 36:2884–2892. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Altuvia Y, Landgraf P, Lithwick G, Elefant
N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T and Margalit H:
Clustering and conservation patterns of human microRNAs. Nucleic
Acids Res. 33:2697–2706. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bentwich I, Avniel A, Karov Y, Aharonov R,
Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al:
Identification of hundreds of conserved and nonconserved human
microRNAs. Nat Genet. 37:766–770. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Veerla S, Lindgren D, Kvist A, Frigyesi A,
Staaf J, Persson H, Liedberg F, Chebil G, Gudjonsson S, Borg A, et
al: MiRNA expression in urothelial carcinomas: Important roles of
miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and
metastasis, and frequent homozygous losses of miR-31. Int J Cancer.
124:2236–2242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu SG, Qin XG, Zhao BS, Qi B, Yao WJ,
Wang TY, Li HC and Wu XN: Differential expression of miRNAs in
esophageal cancer tissue. Oncol Lett. 5:1639–1642. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zheng Q, Sheng Q, Jiang C, Shu J, Chen J,
Nie Z, Lv Z and Zhang Y: MicroRNA-452 promotes tumorigenesis in
hepatocellular carcinoma by targeting cyclin-dependent kinase
inhibitor 1B. Mol Cell Biochem. 389:187–195. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu Q, Gong JP, Li J, Zhong SL, Chen WX,
Zhang JY, Ma TF, Ji H, Lv MM, Zhao JH and Tang JH: Down-regulation
of miRNA-452 is associated with adriamycin-resistance in breast
cancer cells. Asian Pac J Cancer Prev. 15:5137–5142. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kristensen H, Haldrup C, Strand S,
Mundbjerg K, Mortensen MM, Thorsen K, Ostenfeld MS, Wild PJ, Arsov
C, Goering W, et al: Hypermethylation of the GABRE~miR-452~miR-224
promoter in prostate cancer predicts biochemical recurrence after
radical prostatectomy. Clin Cancer Res. 20:2169–2181. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu L, Chen K, Wu J, Shi L, Hu B, Cheng S,
Li M and Song L: Downregulation of miR-452 promotes stem-like
traits and tumorigenicity of gliomas. Clin Cancer Res.
19:3429–3438. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Han L, Pang J, Wang Y, Feng F and
Jiang Q: Expression of microRNA-452 via adenoviral vector inhibits
non-small cell lung cancer cells proliferation and metastasis.
Tumour Biol. 37:8259–8270. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He Z, Xia Y, Pan C, Ma T, Liu B, Wang J,
Chen L and Chen Y: Up-regulation of MiR-452 inhibits metastasis of
non-small cell lung cancer by regulating BMI1. Cell Physiol
Biochem. 37:387–398. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
He Z, Xia Y, Liu B, Qi X, Li Z, Wang J,
Chen L and Chen Y: Down-regulation of miR-452 is associated with
poor prognosis in the non-small-cell lung cancer. J Thorac Dis.
8:894–900. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Merico D, Isserlin R, Stueker O, Emili A
and Bader GD: Enrichment map: A network-based method for gene-set
enrichment visualization and interpretation. PLoS One.
5:e139842010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shannon PT, Grimes M, Kutlu B, Bot JJ and
Galas DJ: RCytoscape: Tools for exploratory network analysis. BMC
Bioinformatics. 14:2172013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Beckers A, Van Peer G, Carter DR, Mets E,
Althoff K, Cheung BB, Schulte JH, Mestdagh P, Vandesompele J,
Marshall GM, et al: MYCN-targeting miRNAs are predominantly
downregulated during MYCN-driven neuroblastoma tumor formation.
Oncotarget. 6:5204–5216. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang J, Chu ES, Chen HY, Man K, Go MY,
Huang XR, Lan HY, Sung JJ and Yu J: microRNA-29b prevents liver
fibrosis by attenuating hepatic stellate cell activation and
inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget.
6:7325–7338. 2015.PubMed/NCBI
|
|
35
|
Zou ZJ, Fan L, Wang L, Xu J, Zhang R, Tian
T, Li JY and Xu W: miR-26a and miR-214 down-regulate expression of
the PTEN gene in chronic lymphocytic leukemia, but not PTEN
mutation or promoter methylation. Oncotarget. 6:1276–1285. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li J, Guan J, Long X, Wang Y and Xiang X:
mir-1-mediated paracrine effect of cancer-associated fibroblasts on
lung cancer cell proliferation and chemoresistance. Oncol Rep.
35:3523–3531. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Peng J, Liu HZ, Zhong J, Deng ZF, Tie CR,
Rao Q, Xu W, You T, Li J, Cai CB, et al: MicroRNA-187 is an
independent prognostic factor in lung cancer and promotes lung
cancer cell invasion via targeting of PTRF. Oncol Rep.
36:2609–2618. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fukazawa T, Guo M, Ishida N, Yamatsuji T,
Takaoka M, Yokota E, Haisa M, Miyake N, Ikeda T, Okui T, et al:
SOX2 suppresses CDKN1A to sustain growth of lung squamous cell
carcinoma. Sci Rep. 6:201132016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cannell IG, Merrick KA, Morandell S, Zhu
CQ, Braun CJ, Grant RA, Cameron ER, Tsao MS, Hemann MT and Yaffe
MB: A pleiotropic RNA-binding protein controls distinct cell cycle
checkpoints to drive resistance of p53-defective tumors to
chemotherapy. Cancer Cell. 28:623–637. 2015. View Article : Google Scholar : PubMed/NCBI
|