Introduction
Histone lysine methylation induced by histone lysine
methyltransferases (HMTs) play a critical role in epigenetic
regulation of biological process, such as gene transcription,
chromatin remodeling, DNA replication and recombination (1,2). On the
N-terminal tail of histone H4, Lys 20 is the only lysine residue
known to be methylated. In mammals, Set8/PR-Set7 catalyzes the
monomethylation of H4K20, whereas Suv4-20h1 and Suv4-20h2 mediate
the dimethylation and trimethylation of H4K20 (3,4).
Disruption of these H4K20 methyltransferases results in genome
instability, thus indicating their important roles in genome
integrity maintenance (4,5). Whereas H4K20me2 is broadly distributed
across the genome, H4K20me3 is enriched in constitutive
heterochromatin, although H4K20me3 is not enriched at the promoters
of inactive genes, as shown by ChIP analyses (3,6–9).
Aberrant histone H4K20 methylation has been
associated with carcinogenesis and cancer progression in various
cancers (10–12). Loss of histone H4K20me3 is considered
a hallmark of cancers, including breast cancer, prostate cancer and
lung cancer (10–12). However, there have been some recently
observed discrepancies in the methylation status of histone H4K20
in some cancers, e.g., breast tumors. There have also been some
conflicting reports in which the levels of H4K20me3 have been found
to be increased (13). Furthermore, a
recent study from a meta-analysis has identified that Suv4-20h1 is
among the 8 (out of 50) dysregulated histone lysine
methyltransferases in breast cancer induced by genetic alterations
(14). Gain or amplification of
Suv4-20h1 has also been observed in most breast cancer cell lines;
therefore, Suv4-20h1 is highly expressed in these cancer cells
compared with non-tumorigenic breast epithelial cells (14). In addition, studies in the TCGA
database have demonstrated that Suv4-20h1 is also amplified in many
types of human cancers, including breast, esophageal, bladder, and
head and neck (15). The expression
profiles of 11 squamous cell carcinoma cell lines have demonstrated
that Suv4-20h1 is significantly highly expressed in these cells
compared with normal keratinocytes (15). However, the functions of Suv4-20h1 in
cancer remain unknown.
In the present study, we sought to characterize the
role of Suv4-20h1 in leukemia K562 cells. We found that Suv4-20h1
accelerates the cell cycle G1/S transition and promotes leukemia
K562 cell growth. The newly identified Suv4-20h1/p21 pathway plays
a key role in the regulation of leukemia K562 cell
proliferation.
Materials and methods
Plasmid construction
The full-length coding sequence (CDS) of the
Suv4-20h1 was amplified from a K562 cDNA template by polymerase
chain reaction (PCR) using primer pairs with EcoR
I/Xho I sites. The PCR products were digested with
EcoR I/Xho I (New England Biolabs, MA, USA), inserted
into the MSCV-3HA-IRES-HA vector, and confirmed by DNA
sequencing.
Cell lines and cell culture
The human chronic myeloid leukemia cell line K562
and its derivatives SCR (stably transfected scrambled shRNA),
Suv4-20h1 KD (stably transfected Suv4-20h1 shRNA), MSCV (stably
transfected MSCV-3HA-IRES-GFP), and MSCV-Suv4-20h1FLC (stably
transfected MSCV-3HA-IRES-GFP+Suv4-20h1 full length CDS) were
established as previously described (16). Cells were maintained in gelatinized
flasks in RPMI 1640 medium supplemented with 10% fetal bovine
serum, 100 U penicillin/100 mg streptomycin (Gibco, Life
Technologies, NY, USA) at 37°C and 5% CO2.
Cell proliferation assay
The cell proliferation index was first analyzed
using a hemocytometer. To accurately measure cell proliferation,
the percentage of S-phase cells in the population was determined by
using a Cell-Light™ EdU flow cytometry assay kit
(Guangzhou RiboBio Co., Ltd., Guangzhou, China), according to the
manufacturer's protocol (http://www.ribobio.com/sitecn/Product.aspx?id=93)
for flow cytometry. EdU (5-ethynyl-2′-deoxyuridine) is a thymidine
analog that is incorporated into DNA during active DNA synthesis.
Newly synthesized DNA and total DNA were detected and quantified by
flow cytometry with EdU and nuclear dyes. The original FACS data
were imported and analyzed in WinMDI 2.9 software to obtain
detailed values for the S-phase cells and all other cell
populations. These values were entered into GraphPad Prism 5
(GraphPad Software, Inc., La Jolla, CA, USA), and then a cell
proliferation histogram was generated.
Apoptosis assay
Cells were collected and washed twice in
phosphate-buffered saline (PBS). Then, the apoptotic and dead cells
were detected with an Annexin V-APC/7-AAD Apoptosis Detection kit
(KeyGEN Biotech, Nanjing, China) according to the manufacturer's
instructions. Data were collected using a BD FACSCalibur Flow
Cytometer. The original FACS data were imported and analyzed in
Flowjo.7.6.1 software (Tree Star, Inc. Ashland, OR, USA) to acquire
detailed values for the different stages of the K562 derivative
cells. These values were entered into GraphPad Prism 5, and then an
apoptosis histogram was generated.
Cell cycle assay
Cells were collected, washed in PBS, and fixed and
permeabilized in 70% ethanol. For determination of DNA content, the
cells were stained with a Cell Cycle Detection Kit (KeyGEN Biotech)
according to the manufacturer's instructions. Data were collected
using a BD FACSCalibur Flow Cytometer. For each sample, 20,000
events were collected, and aggregated cells were gated out. The
original FACS data were imported and analyzed through ModFit LT 3.2
software to acquire the detailed values for the different phases of
the cell cycle. These values were entered into GraphPad Prism 5,
and a cell cycle histogram was generated.
Quantitative RT-PCR
Total RNA was isolated from cells with TRIzol
reagent (Invitrogen, Carlsbad, CA, USA). cDNA was generated using
PrimeScript™ RT Master Mix (Perfect Real Time) kit
(Takara Bio, Otsu, Japan). Quantitative reverse-transcriptase PCR
(qRT-PCR) primers were selected from the PrimerBank (http://pga.mgh.harvard.edu/primerbank/)
or designed online on the Primer3 website: http://bioinfo.ut.ee/primer3-0.4.0/primer3/.
qRT-PCR was performed using FastStart Universal SYBR-Green Master
(Rox) (Roche, South San Francisco, CA) in a final volume of 20 µl
in a Rotorgene 2000 (Corbett Research). Relative quantification was
performed for several potential target genes using GAPDH as an
internal reference. Each reaction was performed in triplicate. The
primers used are listed in Table I.
Student's t-tests were used to derive the significance of the
differences between mean values.
 | Table I.Quantitative RT-PCR primer
sequences. |
Table I.
Quantitative RT-PCR primer
sequences.
| Gene | 5′ Primer | 3′ Primer |
|---|
| CCND1 |
ACCTTCCGCAGTGCTCCTA |
CCCAGCCAAGAAACGGTCC |
| CCND2 |
GGTGGTGCTGGGGAAGTTGAAGTG |
TCGACGGTGGGTACATGGCAAACT |
| CCNB1 |
AGATTGGAGAGGTTGATGTC |
CGATGTGGCATACTTGTTC |
| CCNE1 |
AACTGTGTCAAGTGGATGG |
CTGCTTCTTACCGCTCTG |
| CDK6 |
CTTCATTCACACCGAGTAGT |
TGGACTGGAGCAAGACTT |
| p27 |
GGAGCAATGCGCAGGAATAA |
TGGGGAACCGTCTGAAACAT |
| p21 |
CTGGAGACTCTCAGGGTCGAAA |
GATTAGGGCTTCCTCTTGGAGAA |
| p53 |
GAGGTTGGCTCTGACTGTACC |
TCCGTCCCAGTAGATTACCAC |
| p57 |
ACGATGGAGCGTCTTGTC |
CCTGCTGGAAGTCGTAATC |
| PTEN |
TGGATTCGACTTAGACTTGACCT |
GGTGGGTTATGGTCTTCAAAAGG |
| E2F1 |
AGCTGGACCACCTGATGAAT |
GAGGGGCTTTGATCACCATA |
| SMYD5 |
CCAGCAGCTGCAGCCTCAAAAT |
TGCCGGTGATATTCTGCTCCCCAA |
| Suv4-20h1 |
GAATACTAGCGCCTTTCCTTCG |
GCCCATTCGCCTGAAGTCAA |
| GAPDH |
TGTTGCCATCAATGACCCCTT |
CTCCACGACGTACTCAGCG |
Western blot analysis
Western blot analysis of the cellular extracts was
performed as previously described (16). The specific primary antibodies used in
this study were as follows: Mouse anti-HA monoclonal antibody
(12CA5; Roche), mouse anti-GAPDH monoclonal antibody (sc-47724;
Santa Cruz, Biotechnology, Inc., Santa Cruz, CA), rabbit
anti-p21Waf1/Cip1 antibody (12D1; CST, Beverly, MA,
USA), mouse anti-p53 antibody (1C12; CST), rabbit anti-PTEN
antibody (ab170941; Abcam, Cambridge, MA, USA), rabbit anti-p27
antibody (ab32034; Abcam), mouse anti-E2F1 antibody (05–379;
Millipore Billerica, MA, USA), and rabbit anti-CCND1 antibody
(ab134175; Abcam).
Chromatin immunoprecipitation
(ChIP)
ChIP assays were performed by using standard ChIP
procedures as previously described (17). Chromatin fractions from K562 cell
derivatives were immunoprecipitated with specific antibodies.
Normal rabbit immunoglobulin G (IgG) served as the control. The
antibodies used were as follows: Anti-rabbit IgG (A7028 and A7016;
Beyotime Biotech, Jiangsu, China), anti-HA (12CA5, Roche, South San
Francisco, CA, USA), anti-H4K20me2 (07–367, Millipore), and
anti-H4K20me3 (ab9053, Abcam). Q-PCR was performed using FastStart
Universal SYBR-Green Master mix (Rox) (Roche) in a Rotorgene 2000
instrument (Corbett Research), using immunoprecipitated gDNA. The
detailed values were entered into Origin 7.5, and the relative
enrichment of ChIP DNA was calculated relative to the input DNA.
Each experiment was performed at least twice independently. The
primers used are listed in Table II.
Student's t-tests were used to derive the significance of the
differences between mean values.
 | Table II.ChIP primer sequences. |
Table II.
ChIP primer sequences.
| Locus | 5′ Primer | 3′ Primer |
|---|
| p21 pro |
CATTTGACAACCAGCCCTTT |
TGGGAGGACACAGTAGCAGA |
| E2F1 pro |
CGTTGGCTGTTGGAGATTTT |
TTGCCTCACCCATGACATTA |
Results
Suv4-20h1 promotes K562 cell
proliferation
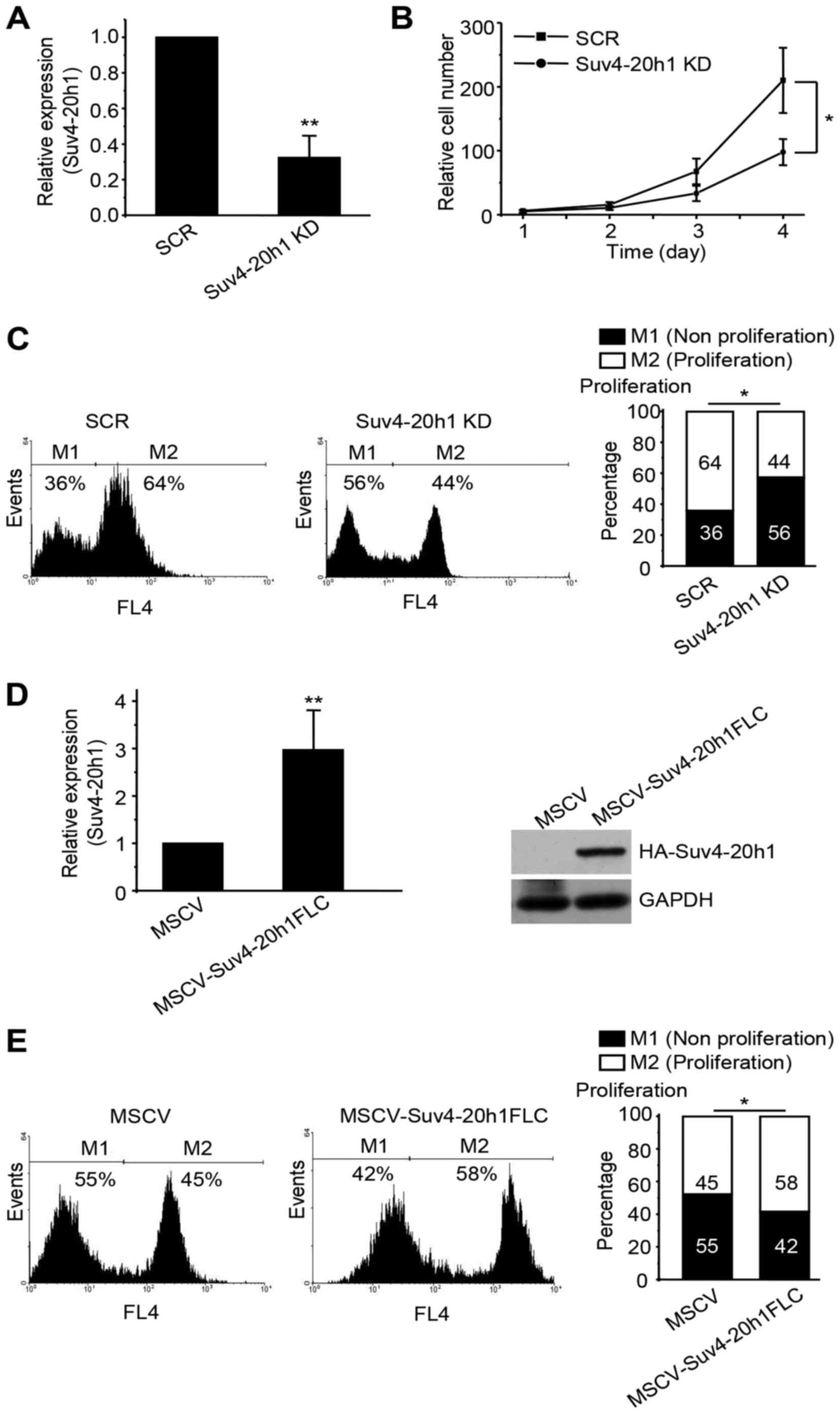
To characterize the function of Suv4-20h1 in K562
cells, we generated a stable Suv4-20h1 knockdown K562 cell line
(Suv4-20h1KD) by using a lentiviral vector containing specific
shRNA. Quantitative real-time PCR confirmed that the expression
levels of Suv4-20h1 were decreased to ~75% of the scrambled control
(SCR) (Fig. 1A). Unfortunately, we
were unable to perform western blot analysis to verify the protein
level of Suv4-20h1, owing to the current lack of commercially
available human-specific anti-Suv4-20h1 antibodies. Consequently,
we observed a significant decrease in the growth rate of the
Suv4-20h1 KD cells compared with the SCR cells (Fig. 1B). To measure the cell proliferation
rate, Cell-Light™ EdU flow cytometry assays were used to
quantify the percentage of S-phase cells in the total cell
population. The results show a separation of proliferating cells
which have incorporated EdU and non-proliferating cells which have
not. We observed fewer proliferative cells in the Suv4-20h1
knockdown cell population than in the SCR cell population (Fig. 1C).
To complement the Suv4-20h1 knockdown cells, we
generated a stable Suv4-20h1 overexpression K562 cell line by using
a retrovirus vector (MSCV-3HA-IRES-GFP) containing the full-length
Suv4-20h1 CDS. Quantitative real-time PCR revealed that levels of
Suv4-20h1 in the MSCV-Suv4-20h1FLC cells were approximately 3-fold
higher than those in the MSCV vector-alone control cells (Fig. 1D). Western blot analysis with anti-HA
antibodies confirmed the presence of the exogenously expressed
HA-tagged Suv4-20h1 (Fig. 1D). We
found that overexpression of Suv4-20h1 significantly increased the
proliferative cell population during the cell cycle (Fig. 1E), thus further confirming that
Suv4-20h1 plays a crucial role in promoting leukemia K562 cell
proliferation.
Knockdown of Suv4-20h1 induces G1/S
cell cycle arrest
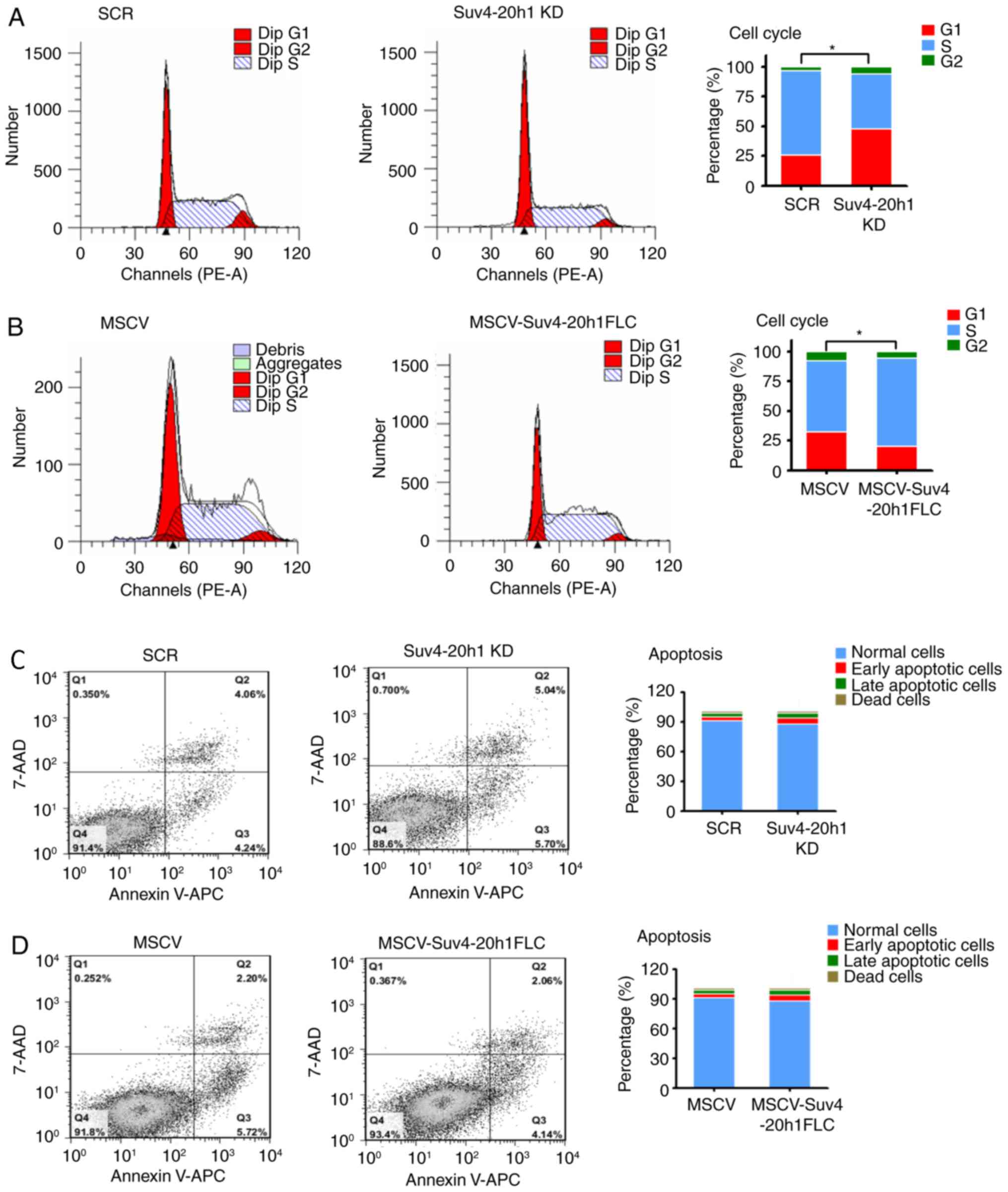
To investigate the effect of Suv4-20h1 on cell cycle
progression, we performed flow cytometric analysis of the cellular
DNA content in K562 cells. We observed more Suv4-20h1 KD cells in
G1 phase compared with SCR cells, which was accompanied by a
decrease in the number of cells in S phase in the cell cycle
(Fig. 2A). In contrast, fewer cells
overexpressing Suv4-20h1FLC were in G1 phase, as compared with the
control cells with MSCV alone, whereas more cells overexpressing
Suv4-20h1FLC were in S phase, as compared with the control cells
with MSCV alone (Fig. 2B). Our
results indicated that knockdown of Suv4-20h1 induces G1-S cell
cycle arrest.
Next, we determined whether Suv4-20h1 plays a role
in cell apoptosis. The total cell apoptosis rate was evaluated by
flow cytometric analysis after double staining with 7-AAD and
Annexin V-APC in which apoptotic cells (positive for Annexin V-APC)
are distinguishable from viable cells (negative for Annexin V-APC
and 7-AAD). We found no changes in the apoptotic cell populations
between Suv4-20h1 KD cells and SCR cells (Fig. 2C). Suv4-20h1-overexpressing cells and
MSCV control cells also displayed a similar pattern of apoptosis
(Fig. 2D). In Fig. 2C and D, there is no difference
(P>0.05) between tested groups and corresponding controls,
including Normal cells, Early apoptotic cells, Late apoptotic cells
and Dead cells. Thus, Suv4-20h1 has no effect on cell apoptosis in
K562 cells, suggesting that Suv4-20h1 promotes cell growth is not
due to less dead cells occurred, rather a faster proliferation of
cells as demonstrated by the EdU assays.
Suv4-20h1 represses p21 expression in
K562 cells
To understand the mechanism underlying the G1/S
phase arrest in Suv4-20h1 knockdown cells, we next examined the
effect of Suv4-20h1 on various key cell cycle regulatory proteins
in the G1 phase of the cell cycle, including CCND1, CCND2, CCNB1,
CCNE1, CDK6, p27, p21, p57, p53, PTEN, E2F1, and SMYD5 (18,19).
Quantitative real-time PCR demonstrated that the expression level
of p21 was most increased in Suv4-20h1 KD cells compared with the
SCR cells (Fig. 3A). In contrast, we
did not observe significant changes in the expression levels of any
other key molecules, such as p53 and PTEN. The effect of Suv4-20h1
on p21 expression was further confirmed at the protein level by
western blotting. The levels of p21 protein were significantly
increased in the Suv4-20h1 knockdown k562 cells (Fig. 3B) compared with controls, thereby
confirming that Suv4-20h1 represses p21 expression.
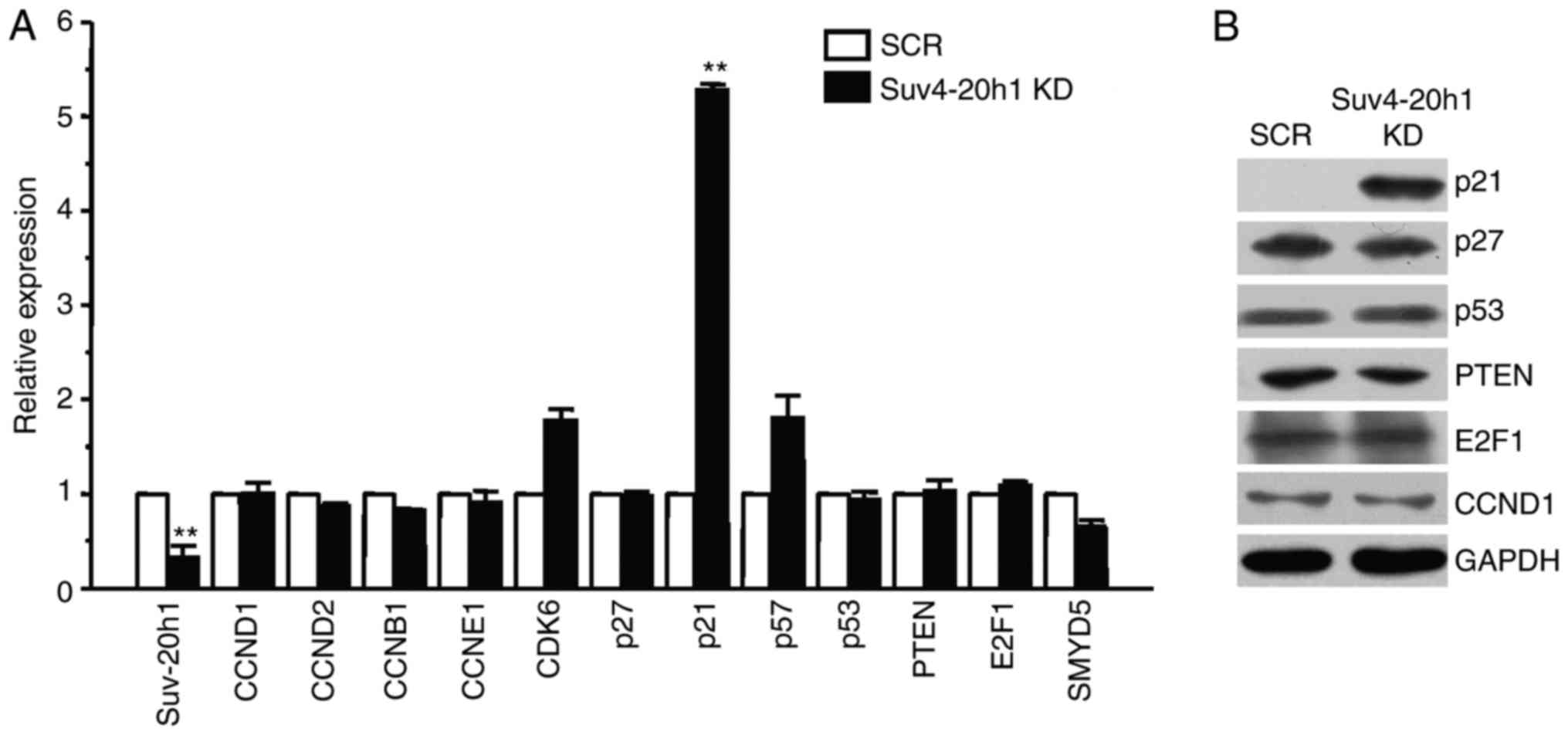 | Figure 3.Suv4-20h1 represses p21 expression in
K562 cells. (A) Suv4-20h1, CCND1, CCND2, CCNB1, CCNE1, CDK6, p27,
p21, p57, p53, PTEN, E2F1 and SMYD5 gene expression analysis by
qRT-PCR of RNA extracted from SCR and Suv4-20h1 KD cells. Data are
normalized to GAPDH mRNA. The results are shown as the mean ± SD
from three independent experiments; **P<0.01 compared with SCR.
(B) Western blot analysis of cellular extracts from SCR and
Suv4-20h1 KD K562 cells, detected with the indicated antibodies.
GAPDH is a loading control. |
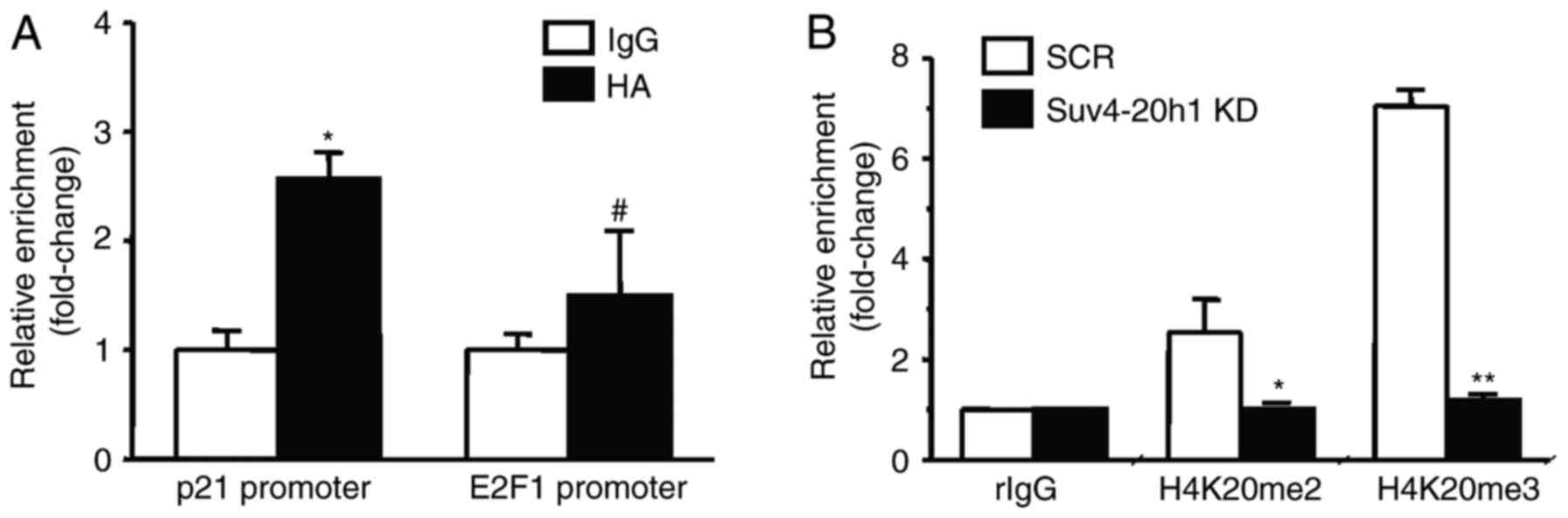
Suv4-20h1 binds the p21 promoter
To examine whether Suv4-20h1 directly regulates p21,
we performed ChIP assays using antibodies against HA in K562 cells
stably overexpressing Suv4-20h1. We found that HA-tagged Suv4-20h1
was significantly enriched at the p21 promoter, as compared with
the IgG control. HA-tagged Suv4-20h1 was not enriched in the E2F1
promoter, a result consistent with our previous E2F1 expression
results (Fig. 4A).
Suv4-20h1 triggers histone H4K20me2/3 activity. To
examine whether Suv4-20h1 affects the histones at the p21 promoter
region, we performed ChIP analysis using antibodies against
H4K20me2 and H4K20me3. We found that enrichment of histones
H4K20me2 and H4K20me3 at the p21 promoter was significantly
decreased in Suv4-20h1 KD cells compared with SCR cells (Fig. 4B). These results indicated that
Suv4-20h1 represses p21 expression via methylation of histone
H4K20.
Discussion
In this study, we sought to investigate the role of
Suv4-20h1 in leukemia K562 cells. We found that knockdown of
Suv4-20h1 resulted in growth inhibition of leukemia K562 cells
through inducing G1 arrest during the cell cycle. A key cell
cycle-related gene, p21, was identified to be a downstream target
of Suv4-20h1 repression. These results demonstrated that Suv4-20h1
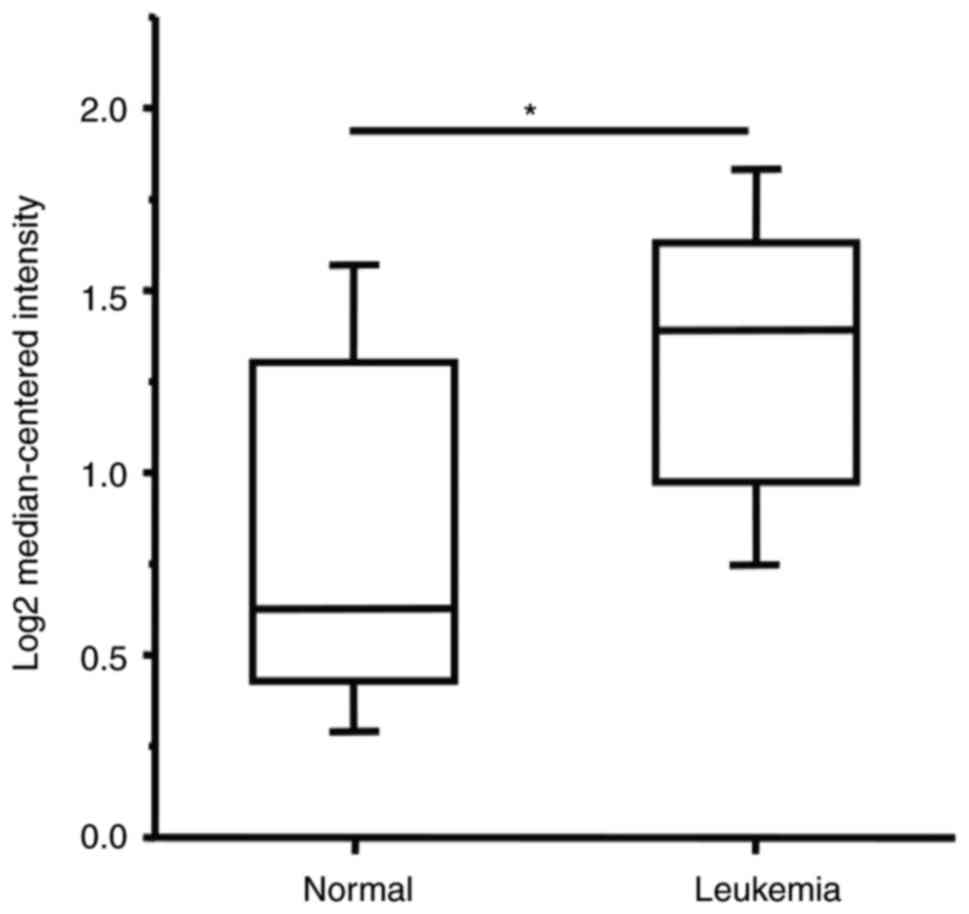
is a potentially oncogenic protein. In fact, we have examined a
deposited high-throughput database ONCOMINE (https://www.oncomine.org) and found that the
expression levels of Suv4-20h1 are indeed significantly higher in
leukemia patients (combined different kinds of leukemia from the
reference) than normal controls (Fig.
5). Similarly, we searched some databases such as TCGA
(https://cancergenome.nih.gov/) and R2
platform (http://r2.amc.nl), but could not find
related data regarding the expression of Suv4-20h1 in the context
of clinical parameters such as Overall Survival (OS), Relapse Free
Survival (RFS).
The cell cycle has been shown to be associated with
the methylation state of histone H4K20 (3,11). The
balance of H4K20 methylation is important in the transition from
cell proliferation to differentiation (3). Among the different methylation states of
H4K20 in the cell cycle, H4K20me1 is the most dynamic, whereas
H4K20me3 is the least abundant and undergoes only modest changes
during the cell cycle (20,21). In proliferating cells, H4K20me2 is the
most abundant form of methylation, which occurs in 80% of all
histone H4 tails (4,20). In mouse embryonic fibroblast cells,
Suv4-20h1 preferentially induces H4K20me2, and this induction may
partially account for the role of Suv4-20h1 in cell proliferation
(4). In fact, in an analysis of mouse
embryonic fibroblasts (MEFs) derived from Suv4-20h1/h2 double
knockout mice (Suv4-20h-dn), a decrease in S-phase cells with a
concomitant increase in G1-phase cells has been shown, as compared
with MEFs derived from wild-type mice, thus indicating a partial
block in the G1/S transition (4).
Consistently with these results, we found that Suv4-20h1 methylates
histone H4 at both H4K20me2 and H4K20me3 in leukemia K562 cells,
and knockdown of Suv4-20h1 caused G1/S stage cell cycle arrest,
thus suggesting that Suv4-20h1 may be one of the major cell cycle
regulators. A similar scenario occurs in head and neck squamous
carcinoma cells in which siRNA-medicated Suv4-20h1 knockdown
induces a significant decrease in the proportion of cells at the S
phase with concomitantly more cells at the G1 phase (15).
Cell cycle progression is precisely regulated by a
series of cell cycle regulators, including cyclins,
cyclin-dependent kinases (CDKs), and CDK inhibitors (CDKIs)
(22–24). p21 is a member of the CDKI family,
which comprises kinase inhibitor proteins or CDK-interacting
proteins, and it inhibits the activity of cyclins at the G1
checkpoint and influences the transition of cells from the G1 phase
to the S phase of the cell cycle (18,24). In
this study, we found that knockdown of Suv4-20h1 significantly
increased p21 expression. Interestingly, we did not observe any
changes in p27 and p57, the two other kinase inhibitor proteins,
after knockdown of Suv4-20h1, thus suggesting the regulatory
specificity of Suv4-20h1 for p21. Therefore, our results indicated
that Suv4-20h1 represses p21 expression and consequently prevents
inhibition of cyclins at the G1 checkpoint, thus promoting the G1/S
transition.
Proper regulation of Suv4-20 h proteins and dynamic
control of H4K20 methylation throughout the cell cycle is of great
importance to maintain cellular homeostasis (4,20). Whereas
loss of H4K20me3 has been regarded as a potential hallmark of human
cancer, these observations have also been inconsistent, with some
reports of increased Suv4-20h1 expression (13–15). There
are three possible explanations for this discrepancy. First, the
precise regulation of histone H4K20 methylation by Set8/PR-Set7,
Suv4-20h1 and Suv4-20h2 may be balanced for proper cell cycle
progression. A Change in one enzyme may not alter the cellular or
local gene histone H4K20 methylation levels in different contexts.
Second, Suv4-20h1 may target other proteins for
methylation/interaction, an effect that may later play a role in
the cell cycle; for example, Suv4-20h1 enhances the phosphorylation
and transcription of ERK1 in cancer cells, thereby promoting cancer
cell proliferation (15). Third, some
discrepancies may be due to differences in the genetic backgrounds
or progressive stages of the cells or specimens. Therefore, careful
analyses are required to determine the relationship between
Suv4-20h1 expression and histone H4K20 methylation status in
different contexts.
In conclusion, our data demonstrated that Suv4-20h1
represses p21 expression to accelerate the G1/S transition during
the cell cycle in human leukemia K562 cells. These data suggested
that Suv4-20h1 plays an important role in cancer cell
proliferation. Given that Suv4-20h1 is frequently overexpressed in
various types of cancers, inhibition of Suv4-20h1 may be an
alternative therapeutic strategy for cancer patients exhibiting
this overexpression.
Acknowledgements
The authors would like to thank members of the Zhao
laboratory for helpful discussions. The present study was supported
by the National Natural Science Foundation of China NSFC [grant
nos. 31470750 and 31270811 (to Q. Z.), grant no. 2015M571736 (to J.
J.) and grant no. 2016M590442 (to M. L.)].
References
|
1
|
Greer EL and Shi Y: Histone methylation: A
dynamic mark in health, disease and inheritance. Nat Rev Genet.
13:343–357. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chi P, Allis CD and Wang GG: Covalent
histone modifications-miswritten, misinterpreted and mis-erased in
human cancers. Nat Rev Genet. 10:457–469. 2010.
|
|
3
|
Jorgensen S, Schotta G and Sørensen CS:
Histone H4 lysine 20 methylation: Key player in epigenetic
regulation of genomic integrity. Nat Rev Genet. 41:2797–2806.
2013.
|
|
4
|
Schotta G, Sengupta R, Kubicek S, Malin S,
Kauer M, Callén E, Celeste A, Pagani M, Opravil S, De La
Rosa-Velazquez IA, et al: A chromatin-wide transition to H4K20
monomethylation impairs genome integrity and programmed DNA
rearrangements in the mouse. Genes Dev. 22:2048–2061. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schotta G, Lachner M, Sarma K, Ebert A,
Sengupta R, Reuter G, Reinberg D and Jenuwein T: A silencing
pathway to induce H3-K9 and H4-K20 trimethylation at constitutive
heterochromatin. Genes Dev. 18:1251–1262. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kohlmaier A, Savarese F, Lachner M,
Martens J, Jenuwein T and Wutz A: A chromosomal memory triggered by
Xist regulates histone methylation in X inactivation. PLoS Biol.
2:E1712004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sims JK, Houston SI, Magazinnik T and Rice
JC: A trans-tail histone code defined by monomethylated H4 Lys-20
and H3 Lys-9 demarcates distinct regions of silent chromatin. J
Biol Chem. 281:12760–12766. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Barski A, Cuddapah S, Cui K, Roh TY,
Schones DE, Wang Z, Wei G, Chepelev I and Zhao K: High-resolution
profiling of histone methylations in the human genome. Cell.
129:823–837. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mikkelsen TS, Ku M, Jaffe DB, Issac B,
Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP,
et al: Genome-wide maps of chromatin state in pluripotent and
lineage-committed cells. Nature. 448:553–560. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fraga MF, Ballestar E, Villar-Garea A,
Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S,
Petrie K, et al: Loss of acetylation at Lys16 and trimethylation at
Lys20 of histone H4 is a common hallmark of human cancer. Nat
Genet. 37:391–400. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Balakrishnan L and Milavetz B: Decoding
the histone H4 lysine 20 methylation mark. Crit Rev Biochem Mol
Biol. 45:440–452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Van Den Broeck A, Brambilla E,
Moro-Sibilot D, Lantuejoul S, Brambilla C, Eymin B, Khochbin S and
Gazzeri S: Loss of histone H4K20 trimethylation occurs in
preneoplasia and influences prognosis of non-small cell lung
cancer. Clin Cancer Res. 14:7237–7245. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Elsheikh SE, Green AR, Rakha EA, Powe DG,
Ahmed RA, Collins HM, Soria D, Garibaldi JM, Paish CE, Ammar AA, et
al: Global histone modifications in breast cancer correlate with
tumor phenotypes, prognostic factors and patient outcome. Cancer
Res. 69:3802–3809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu L, Kimball S, Liu H, Holowatyj A and
Yang ZQ: Genetic alterations of histone lysine methyltransferases
and their significance in breast cancer. Oncotarget. 6:2466–2482.
2015.PubMed/NCBI
|
|
15
|
Vougiouklakis T, Sone K, Saloura V, Cho
HS, Suzuki T, Dohmae N, Alachkar H, Nakamura Y and Hamamoto R:
SUV420H1 enhances the phosphorylation and transcription of ERK1 in
cancer cells. Oncotarget. 6:43162–43171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rank G, Cerruti L, Simpson RJ, Moritz RL,
Jane SM and Zhao Q: Identification of a PRMT5-dependent repressor
complex linked to silencing of human fetal globin gene expression.
Blood. 116:1585–1592. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao Q, Rank G, Tan YT, Li H, Moritz RL,
Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, et al:
PRMT5-mediated methylation of histone H4R3 recruits DNMT3A,
coupling histone and DNA methylation in gene silencing. Nat Struct
Mol Biol. 16:304–311. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kreis NN, Louwen F and Yuan J: Less
understood issues: p21(Cip1) in mitosis and its therapeutic
potential. Oncogene. 34:1758–1767. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tanaka T and Iino M: Knockdown of Sec8
promotes cell-cycle arrest at G1/S phase by inducing p21 via
control of FOXO proteins. FEBS J. 281:1068–1084. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pesavento JJ, Yang H, Kelleher NL and
Mizzen CA: Certain and progressive methylation of histone H4 at
lysine 20 during the cell cycle. Mol Cell Biol. 28:468–486. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tsang LW, Hu N and Underhill DA:
Comparative analyses of SUV420H1 isoforms and SUV420H2 reveal
differences in their cellular localization and effects on myogenic
differentiation. PLoS One. 5:e144472010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lim S and Kaldis P: Cdks, cyclins and
CKIs: Roles beyond cell cycle regulation. Development.
140:3079–3093. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Malumbres M and Barbacid M: Cell cycle,
CDKs and cancer: A changing paradigm. Nat Rev Cancer. 9:153–166.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bertoli C, Skotheim JM and de Bruin RA:
Control of cell cycle transcription during G1 and S phases. Nat Rev
Mol Cell Biol. 14:518–528. 2013. View
Article : Google Scholar : PubMed/NCBI
|