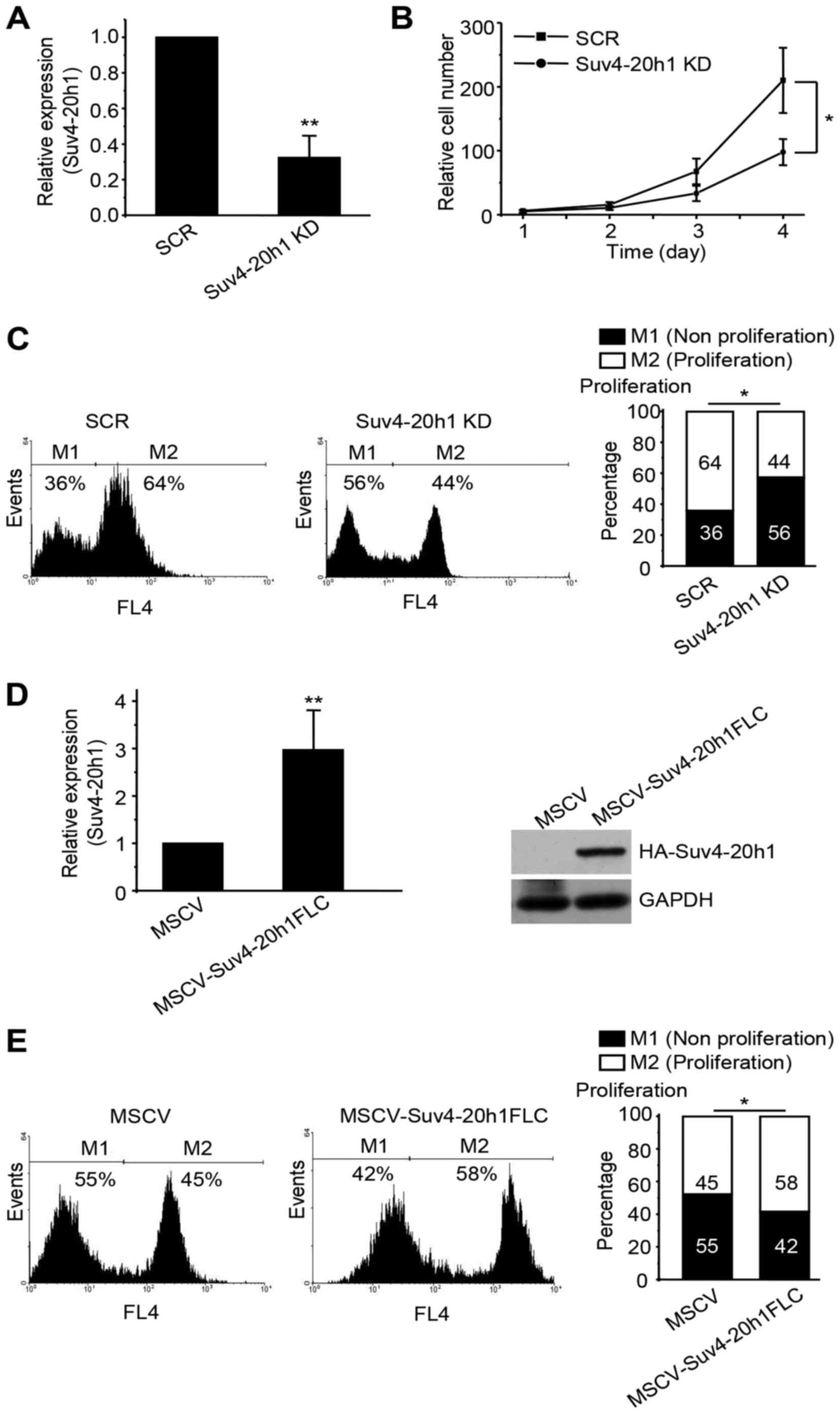
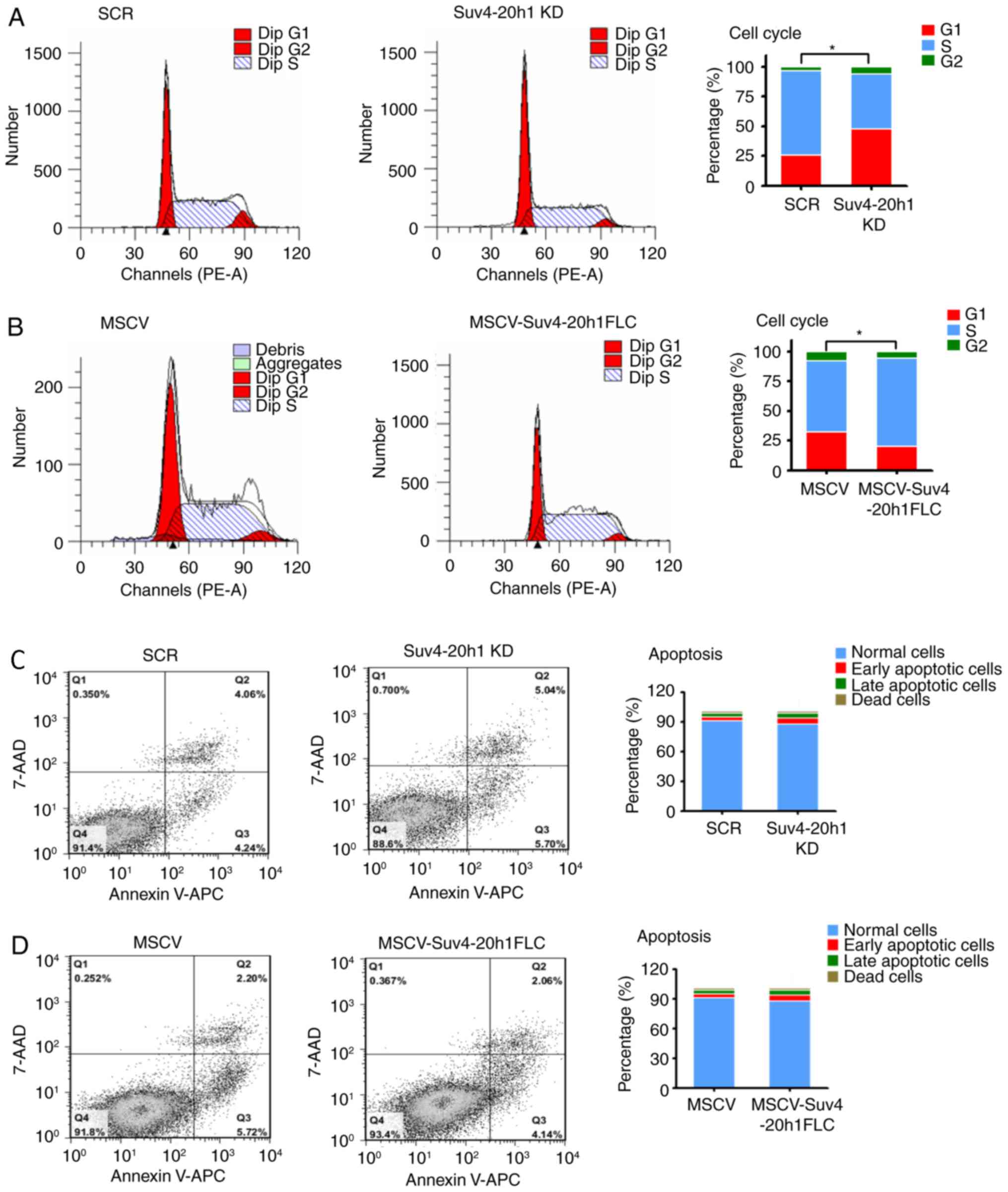
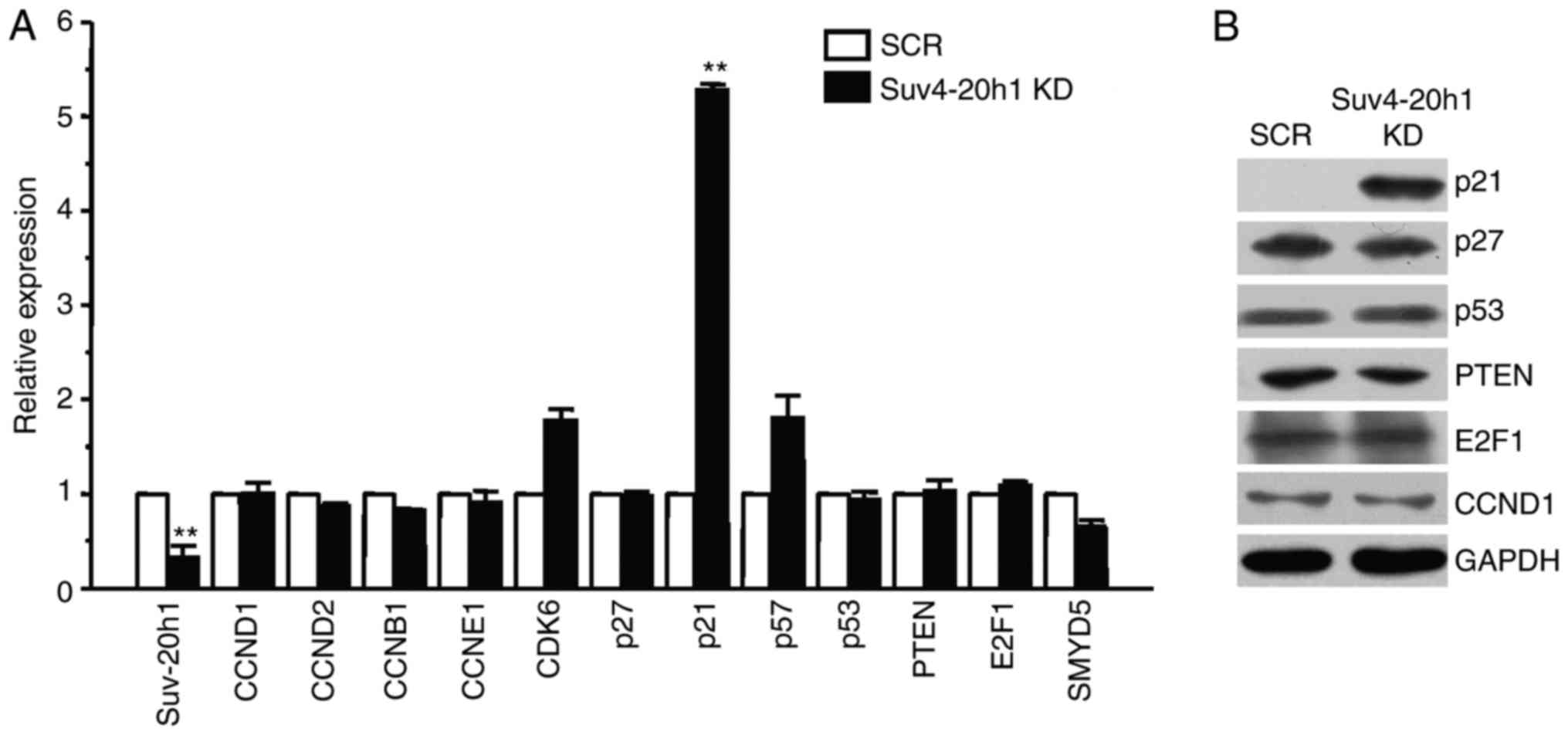
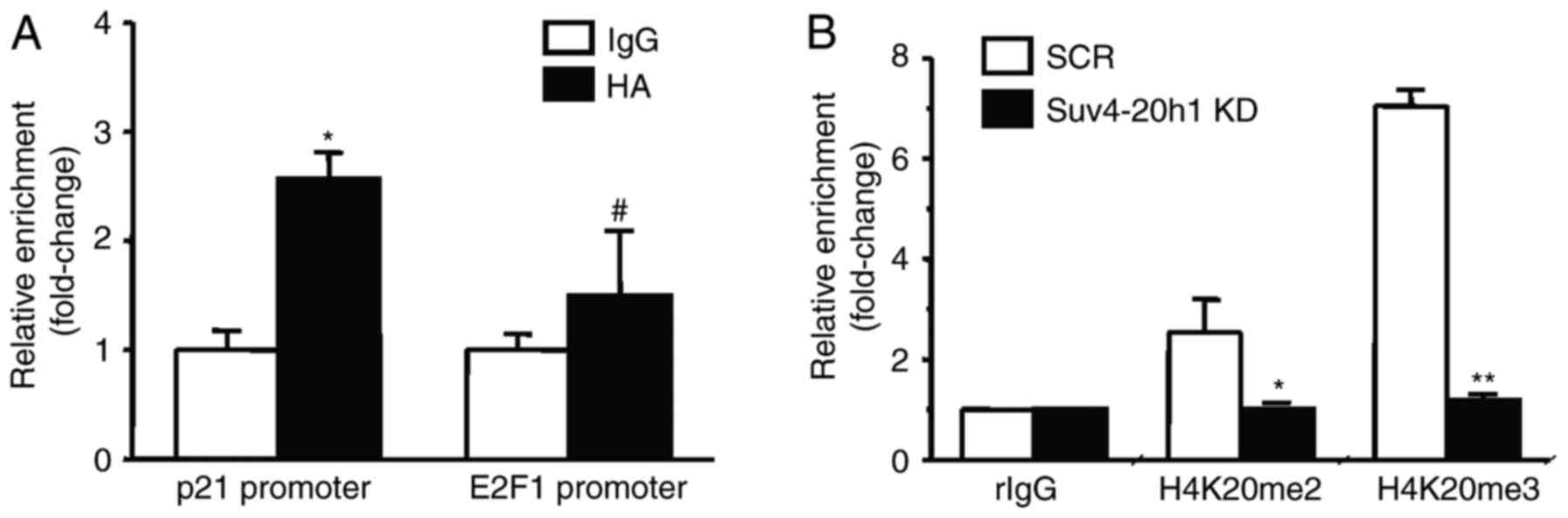
|
1
|
Greer EL and Shi Y: Histone methylation: A
dynamic mark in health, disease and inheritance. Nat Rev Genet.
13:343–357. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chi P, Allis CD and Wang GG: Covalent
histone modifications-miswritten, misinterpreted and mis-erased in
human cancers. Nat Rev Genet. 10:457–469. 2010.
|
|
3
|
Jorgensen S, Schotta G and Sørensen CS:
Histone H4 lysine 20 methylation: Key player in epigenetic
regulation of genomic integrity. Nat Rev Genet. 41:2797–2806.
2013.
|
|
4
|
Schotta G, Sengupta R, Kubicek S, Malin S,
Kauer M, Callén E, Celeste A, Pagani M, Opravil S, De La
Rosa-Velazquez IA, et al: A chromatin-wide transition to H4K20
monomethylation impairs genome integrity and programmed DNA
rearrangements in the mouse. Genes Dev. 22:2048–2061. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schotta G, Lachner M, Sarma K, Ebert A,
Sengupta R, Reuter G, Reinberg D and Jenuwein T: A silencing
pathway to induce H3-K9 and H4-K20 trimethylation at constitutive
heterochromatin. Genes Dev. 18:1251–1262. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kohlmaier A, Savarese F, Lachner M,
Martens J, Jenuwein T and Wutz A: A chromosomal memory triggered by
Xist regulates histone methylation in X inactivation. PLoS Biol.
2:E1712004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sims JK, Houston SI, Magazinnik T and Rice
JC: A trans-tail histone code defined by monomethylated H4 Lys-20
and H3 Lys-9 demarcates distinct regions of silent chromatin. J
Biol Chem. 281:12760–12766. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Barski A, Cuddapah S, Cui K, Roh TY,
Schones DE, Wang Z, Wei G, Chepelev I and Zhao K: High-resolution
profiling of histone methylations in the human genome. Cell.
129:823–837. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mikkelsen TS, Ku M, Jaffe DB, Issac B,
Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP,
et al: Genome-wide maps of chromatin state in pluripotent and
lineage-committed cells. Nature. 448:553–560. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fraga MF, Ballestar E, Villar-Garea A,
Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S,
Petrie K, et al: Loss of acetylation at Lys16 and trimethylation at
Lys20 of histone H4 is a common hallmark of human cancer. Nat
Genet. 37:391–400. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Balakrishnan L and Milavetz B: Decoding
the histone H4 lysine 20 methylation mark. Crit Rev Biochem Mol
Biol. 45:440–452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Van Den Broeck A, Brambilla E,
Moro-Sibilot D, Lantuejoul S, Brambilla C, Eymin B, Khochbin S and
Gazzeri S: Loss of histone H4K20 trimethylation occurs in
preneoplasia and influences prognosis of non-small cell lung
cancer. Clin Cancer Res. 14:7237–7245. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Elsheikh SE, Green AR, Rakha EA, Powe DG,
Ahmed RA, Collins HM, Soria D, Garibaldi JM, Paish CE, Ammar AA, et
al: Global histone modifications in breast cancer correlate with
tumor phenotypes, prognostic factors and patient outcome. Cancer
Res. 69:3802–3809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu L, Kimball S, Liu H, Holowatyj A and
Yang ZQ: Genetic alterations of histone lysine methyltransferases
and their significance in breast cancer. Oncotarget. 6:2466–2482.
2015.PubMed/NCBI
|
|
15
|
Vougiouklakis T, Sone K, Saloura V, Cho
HS, Suzuki T, Dohmae N, Alachkar H, Nakamura Y and Hamamoto R:
SUV420H1 enhances the phosphorylation and transcription of ERK1 in
cancer cells. Oncotarget. 6:43162–43171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rank G, Cerruti L, Simpson RJ, Moritz RL,
Jane SM and Zhao Q: Identification of a PRMT5-dependent repressor
complex linked to silencing of human fetal globin gene expression.
Blood. 116:1585–1592. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao Q, Rank G, Tan YT, Li H, Moritz RL,
Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, et al:
PRMT5-mediated methylation of histone H4R3 recruits DNMT3A,
coupling histone and DNA methylation in gene silencing. Nat Struct
Mol Biol. 16:304–311. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kreis NN, Louwen F and Yuan J: Less
understood issues: p21(Cip1) in mitosis and its therapeutic
potential. Oncogene. 34:1758–1767. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tanaka T and Iino M: Knockdown of Sec8
promotes cell-cycle arrest at G1/S phase by inducing p21 via
control of FOXO proteins. FEBS J. 281:1068–1084. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pesavento JJ, Yang H, Kelleher NL and
Mizzen CA: Certain and progressive methylation of histone H4 at
lysine 20 during the cell cycle. Mol Cell Biol. 28:468–486. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tsang LW, Hu N and Underhill DA:
Comparative analyses of SUV420H1 isoforms and SUV420H2 reveal
differences in their cellular localization and effects on myogenic
differentiation. PLoS One. 5:e144472010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lim S and Kaldis P: Cdks, cyclins and
CKIs: Roles beyond cell cycle regulation. Development.
140:3079–3093. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Malumbres M and Barbacid M: Cell cycle,
CDKs and cancer: A changing paradigm. Nat Rev Cancer. 9:153–166.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bertoli C, Skotheim JM and de Bruin RA:
Control of cell cycle transcription during G1 and S phases. Nat Rev
Mol Cell Biol. 14:518–528. 2013. View
Article : Google Scholar : PubMed/NCBI
|