Introduction
Epithelial ovarian cancer (EOC) is the most common
type of invasive ovarian cancer, and a leading cause of
cancer-associated mortalities in females globally (1,2). In China,
the incidence rate of ovarian cancer was 7.91/100,000 and the
age-adjusted incidence rate was 5.35/100,000 between 1999 and 2010
(3). The incidence of EOC increases
rapidly following menopause (4).
Although a number of factors are involved the in the molecular
mechanism behind EOC, these risk factors are not well defined and
the pathogenesis of EOC remains unclear (5). The progress in the treatment of EOC is
not optimistic, the prognosis is poor and the 5-year relative
survival rate is ~30% (6).
In recent years the functions of microRNAs
(miRNAs/miRs) in the cancer have begun to be revealed (7–12), and a
number of miRNAs have been identified to be involved in the
pathogenesis of EOC (13–19). miRNAs are evolutionarily conserved,
small non-coding RNAs that regulate the expression of genes and
translocation of proteins (20).
The functions of miR-545 have been studied in
several types of cancer. In pancreatic ductal adenocarcinoma,
miR-545 inhibited cellular proliferation and was associated with a
decreased survival rate in patients (21). In lung cancer, miR-545 suppressed
cellular proliferation via the inhibition of cyclin D1 and
cyclin-dependent kinase 4 expression (22). However, the function of miR-545 in EOC
remains unknown.
In the present study, the expression level of
miR-545 was analyzed in EOC tissues and the in vitro
functions of miR-545 were investigated. Subsequently, the potential
miR-545-target genes were assessed.
Materials and methods
Patients and tissues
A total of 27 EOC tissues and matched normal
adjacent tissues were collected from 27 subjects (the mean age was
59.9 years, and the range was 36–45 years) who underwent surgery
for the treatment of EOC at the West China Women's and Children's
Hospital of Sichuan University (Chengdu, China). The
histopathological diagnosis of EOC was provided and confirmed by
the senior pathologist of the West China Second University
Hospital, Sichuan University (Chengdu, Sichuan 610041). The
survival status of patients was recorded and used for Kaplan-Meier
survival analysis. Patient information is summarized in Table I. The present study was approved by
Ethic Committee of Sichuan University and each patient provided
written informed consent.
 | Table I.Characteristics of patients with
ovarian cancer. |
Table I.
Characteristics of patients with
ovarian cancer.
| No. | Age, years | Type | Stage | Follow-up time,
months | Status |
|---|
| 1 | 45 | Epithelial ovarian
cancer/serous | IV | 34 | Deceased |
| 2 | 54 | Epithelial ovarian
cancer/serous | IV | 44 | Deceased |
| 3 | 46 | Epithelial ovarian
cancer/serous | III | 55 | Deceased |
| 4 | 56 | Epithelial ovarian
cancer/serous | III | 66 | Survived |
| 5 | 48 | Epithelial ovarian
cancer/serous | IV | 66 | Survived |
| 6 | 36 | Epithelial ovarian
cancer/serous | III | 66 | Survived |
| 7 | 57 | Epithelial ovarian
cancer/serous | III | 66 | Survived |
| 8 | 45 | Epithelial ovarian
cancer/serous | IV | 66 | Survived |
| 9 | 64 | Epithelial ovarian
cancer/serous | IV | 66 | Survived |
| 10 | 75 | Epithelial ovarian
cancer/serous | IV | 66 | Survived |
| 11 | 56 | Epithelial ovarian
cancer/serous | IV | 66 | Survived |
| 12 | 74 | Epithelial ovarian
cancer/serous | IV | 66 | Survived |
| 13 | 69 | Epithelial ovarian
cancer/serous | IV | 66 | Survived |
| 14 | 79 | Epithelial ovarian
cancer/serous | IV | 4 | Deceased |
| 15 | 75 | Epithelial ovarian
cancer/serous | IV | 7 | Deceased |
| 16 | 67 | Epithelial ovarian
cancer/serous | IV | 11 | Deceased |
| 17 | 65 | Epithelial ovarian
cancer/serous | III | 13 | Deceased |
| 18 | 69 | Epithelial ovarian
cancer/serous | IV | 14 | Deceased |
| 19 | 73 | Epithelial ovarian
cancer/serous | IV | 19 | Deceased |
| 20 | 65 | Epithelial ovarian
cancer/serous | IV | 22 | Deceased |
| 21 | 59 | Epithelial ovarian
cancer/serous | IV | 27 | Deceased |
| 22 | 57 | Epithelial ovarian
cancer/serous | III | 66 | Deceased |
| 23 | 63 | Epithelial ovarian
cancer/serous | IV | 66 | Survived |
| 24 | 67 | Epithelial ovarian
cancer/serous | IV | 66 | Survived |
| 25 | 54 | Epithelial ovarian
cancer/serous | III | 66 | Survived |
| 26 | 51 | Epithelial ovarian
cancer/serous | IV | 66 | Survived |
| 27 | 49 | Epithelial ovarian
cancer/serous | IV | 66 | Survived |
Cell lines
The EOC SKOV3 and HO8910 cell lines and the normal
immortalized human ovarian surface epithelial cells IOSE29 were
acquired from the Type Culture Collection of the Chinese Academy of
Sciences (Shanghai, China). Cells were cultured in RPMI-1640 medium
with 10% fetal bovine serum (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) (23).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Following cell and tissue isolation, total RNA was
purified from EOC tissues, IOSE29, SKOV3, HO8910 cell lines
(treated or control group) using TRIzol reagent (Life Sciences;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. The extracted RNA was dissolved in DEPC-treated
ddH2O and subjected to DNAse I treatment (catalog no,
EN0521; Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was
reverse-transcribed to cDNA by using All-in-One™ miRNA
First-Strand cDNA Synthesis kit (catalog no. AMRT-0020; Biocompare,
South San Francisco, CA, USA). The PCR process was completed by
SYBR® Green PCR Master Mix (catalog no. 4364344; Thermo
Fisher Scientific, Inc.). The primers were synthesized and tested
by the Chengdu Haoyi Biotechnology Co., Ltd (http://haoyibio.net) (Chengdu, China). The following
primers were used for the analysis: miR-545, 5S rRNA forward
primer, 5′-ACGGCCATACCACCCTGAAC-3′, reverse primer,
5′-GGCGGTCTCCCATCCAAGTA-3′; U6 forward primer,
5′-CTCGCTTCGGCAGCACA-3′, reverse primer,
5′-AACGCTTCACGAATTTGCGT-3′. Data analysis were performed using the
the 2−ΔΔCq method of normalization (24). U6 was used for the normalization of
miR-545 (10,25–27).
miR-545 mimics and anti-sense
oligonucleotides transfection (ASO)
The miR-545 mimics (5′-UCAGCAAACAUUUAUUGUGUGC-3′),
miR-545 ASO (5′-GCACACAAUAAAUGUUUGCUGA-3′) and negative controls
(5′-UGGGCGUAUAGACGUGUUACAC-3′) were purchased from Dharmacon (GE
Healthcare Life Sciences, Little Chalfont, UK). Cells were seeded
at 2×105 per well in 6-well plates for further
investigation. These cells were transfected with miR-545 mimics,
miR-545 ASO or miR-545 mimics using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. After 24 h, the transfected cells were used for further
experiments.
Cell proliferation assay
The cellular proliferation was assessed using an MTT
assay. SKOV3 and HO8910 cells were seeded into 96-well plates at a
density of 5×105 cells/well. MTT reagent was added into
the medium at a final concentration of 0.1 mg/ml and the plate was
incubated at 37°C for 3 h. Following incubation, Dimethyl sulfoxide
(100 µl) was added at 37°C for 15 min. Optical density was measured
using a microplate reader at 570 nm.
Cell apoptosis analysis
Annexin V-fluorescein isothiocyanate (FITC)
Staining/Detection kit (ab14085, Abcam, Cambridge, UK) was used.
The binding buffer were prepared according to the manufacturer's
protocol. Treated SKOV3 and HO8910 cells were suspended
(5×105 cells/ml) in 1xbinding buffer. Annexin V-FITC (5
µl) was added and incubated for 15 min at room temperature.
Subsequently, propidium iodide (5 µl) (ab14083, Abcam) was added
for 5 min at room temperature, samples were analyzed with a flow
cytometer using the 488 nm excitation line (Argon-ion laser or
solid state laser) and emission detected at 530 nm (green, FITC)
and 575–610 nm (orange, PI). The data was analyzed by BD LSR II
software (v.1.1.0, Franklin, NJ, USA).
Dual luciferase reporter assays
SKOV3 cells were seeded at 1×105 per well
and were serum-starved for 6 h prior to transfection. The RAC-γ
serine/threonine-protein kinase (AKT3) 3′-UTR reporter plasmid (500
ng) and the pGL3-control (100 ng) (Promega Corporation, Madison,
WI, USA) were co-transfected into cells using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). Cells were harvested
and luciferase activities were analyzed after 24 h using the
Dual-Luciferase Reporter Assay system (Promega Corporation).
Mutants of AKT3 3′-UTR were generated using a Site-Directed
Mutagenesis kit (Promega Corporation). The data was normalized by
Renilla luciferase activity. The value in miR-NC group was
arbitrarily defined as 100%.
Prediction of the possible targets of
miR-545
Targetscan software (http://www.targetscan.org/) was used to predict the
possible targets of miR-545 by searching miR-545. TargetScan is
online software, that predicts biological targets of miRNA by
searching for the presence of conserved 8mer, 7mer, and 6mer sites
that match the seed region of each miRNA (28).
Western blot analysis
The lysates were prepared with lysis buffer (M-PER™
Mammalian Protein Extraction Reagent, Thermo Fisher Scientific,
Inc., Waltham, MA, USA) containing protease inhibitors (Pierce
Protease Inhibitor, A32963, Thermo Fisher Scientific, Inc.) and
then were cleared by centrifugation. The protein concentration was
assayed by Pierce™ Rapid Gold BCA Protein Assay Kit
(Thermo Fisher Scientific, Inc.). Total proteins (30 µg) were
resolved using SDS-PAGE (10%) and transferred onto polyvinylidene
difluoride membranes (Merck KGaA, Darmstadt, Germany). The
membranes were immersed with 5% non-fat dry milk (in Blocking
Buffer, Thermo Fisher Scientific, Inc.) for 15 min at room
temperature. Membranes were washed with PBS for 15 min and then
incubated with primary antibodies overnight in 4°C, washed by PBS
for three times. Then the membranes were incubated with secondary
antibodies for 2 h at room temperature. Western blot Signal was
developed by Pierce™ DAB Substrate Kit (Thermo Fisher, Scientific,
Inc.), then the blotting images were captured using
CL-XPosure™ film 5×7 inch (Thermo Fisher Scientific,
Inc.). The following antibodies were used as follows: AKT3 (catalog
no. sc-56878, 1:1,000 dilution; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA), GAPDH (catalog no. sc69778, 1:3,000 dilution;
Santa Cruz Biotechnology, Inc.), goat anti-rabbit (catalog no.
sc-2004, 1:10,000 dilution; Santa Cruz Biotechnology, Inc.).
Statistical analysis
Data are presented as mean ± standard deviation.
Statistical analysis was performed using SPSS software (version 10;
SPSS, Inc., Chicago, IL, USA). The difference between two groups
was analyzed using a two-tailed Student's t-test. One-way analysis
of variance (ANOVA) was used to analyze the difference between
three groups; Fisher's least significant difference test was
performed following ANOVA. Analysis of the survival rate of
patients with EOC was performed via Kaplan-Meier survival curves
and a log-rank test was performed to confirm the significance.
P<0.05 was considered to indicate a statistically significant
difference.
Results
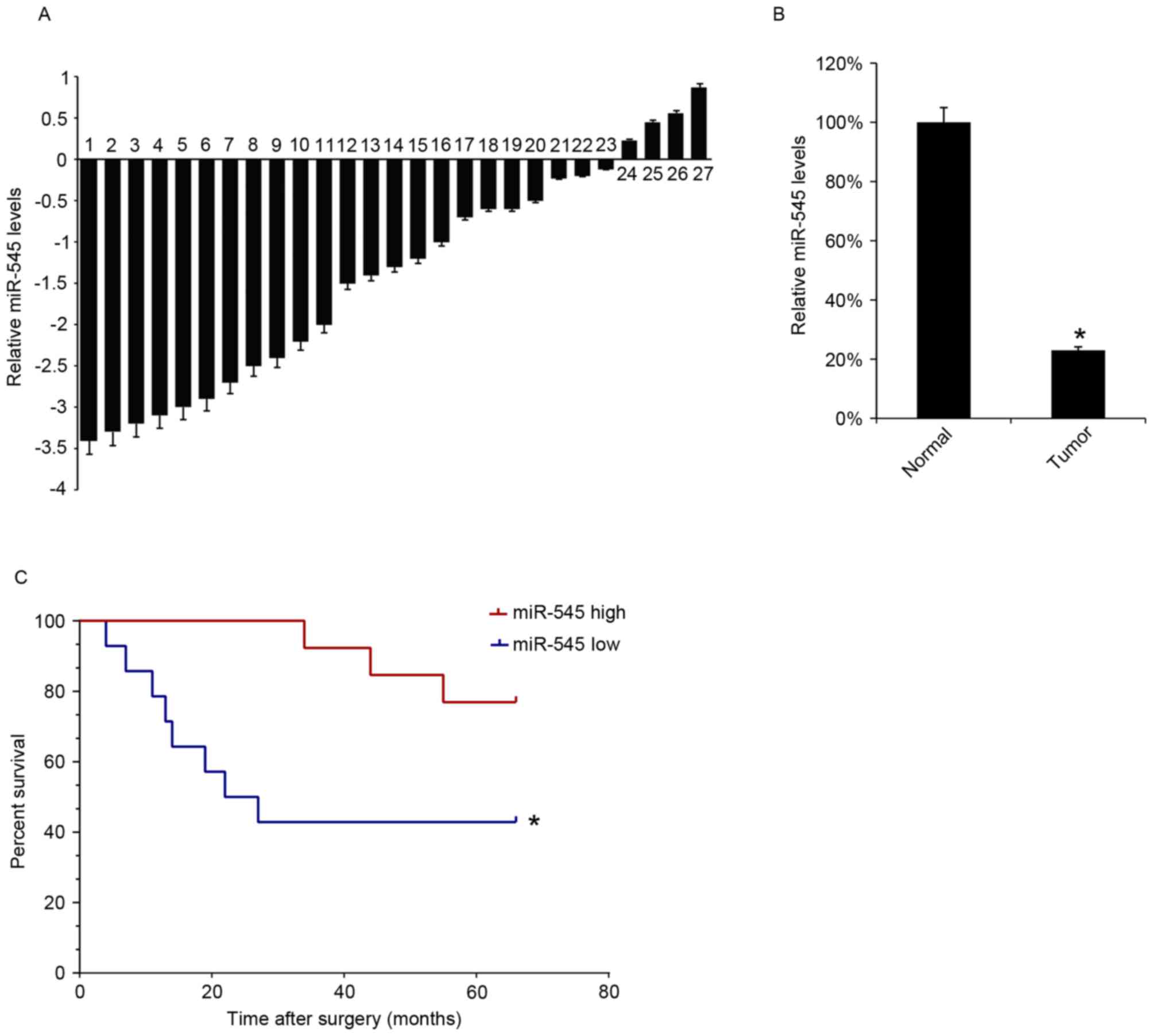
miR-545 expression is decreased in EOC
tissues and low level of miR-545 is associated with low survival
rate of patients with EOC
The miR-545 levels in 27 EOC tissues samples were
analyzed using RT-qPCR. The levels of miR-545 expression were
decreased in tumor tissues compared with matched-adjacent normal
tissues in 23 specimens (Fig. 1A).
The mean expression of miR-545 in the 27 EOC tissues samples was
significantly lower than that in the adjacent normal samples
(Fig. 1B). The survival rate of the
patients for 66 months following surgical treatment was also
assessed, and the 27 patients with EOC were divided into two groups
according to the median value of the miR-545 expression in EOC
tissues. Kaplan-Meyer survival analysis revealed that patients
expressing increased levels of miR-545 in EOC tissues were
associated with increased survival rates (Fig. 1C).
Overexpression of miR-545 inhibits
cells growth and promotes apoptosis
Next the function of miR-545 in the EOC cell lines
SKOV3 and HO8910 was tested. It was identified that miR-545 was
expressed at decreased levels in SKOV3 and HO8910 cells compared
with normal ovarian tissue and the normal immortalized human
ovarian surface epithelial cells IOSE29 (Fig. 2A). Next the miR-545 levels in SKOV3
and HO8910 cells were determined following transfection with
miR-545 mimics; miR-545 mimics upregulated the miR-545 levels in
the two cell lines at 48 and 72 h after transfection (Fig. 2B). Cellular proliferation and
apoptosis were assessed using MTT analysis at 48 h after
transfection with miR-545 mimics or the negative control.
Overexpression of miR-545 was revealed to inhibit cellular
proliferation (Fig. 2C). At 48 h
after transfection, the rates of cell apoptosis were assessed using
flow cytometry, and it was identified that miR-545 increased the
apoptosis rate in SKOV3 and HO8910 (Fig.
2D).
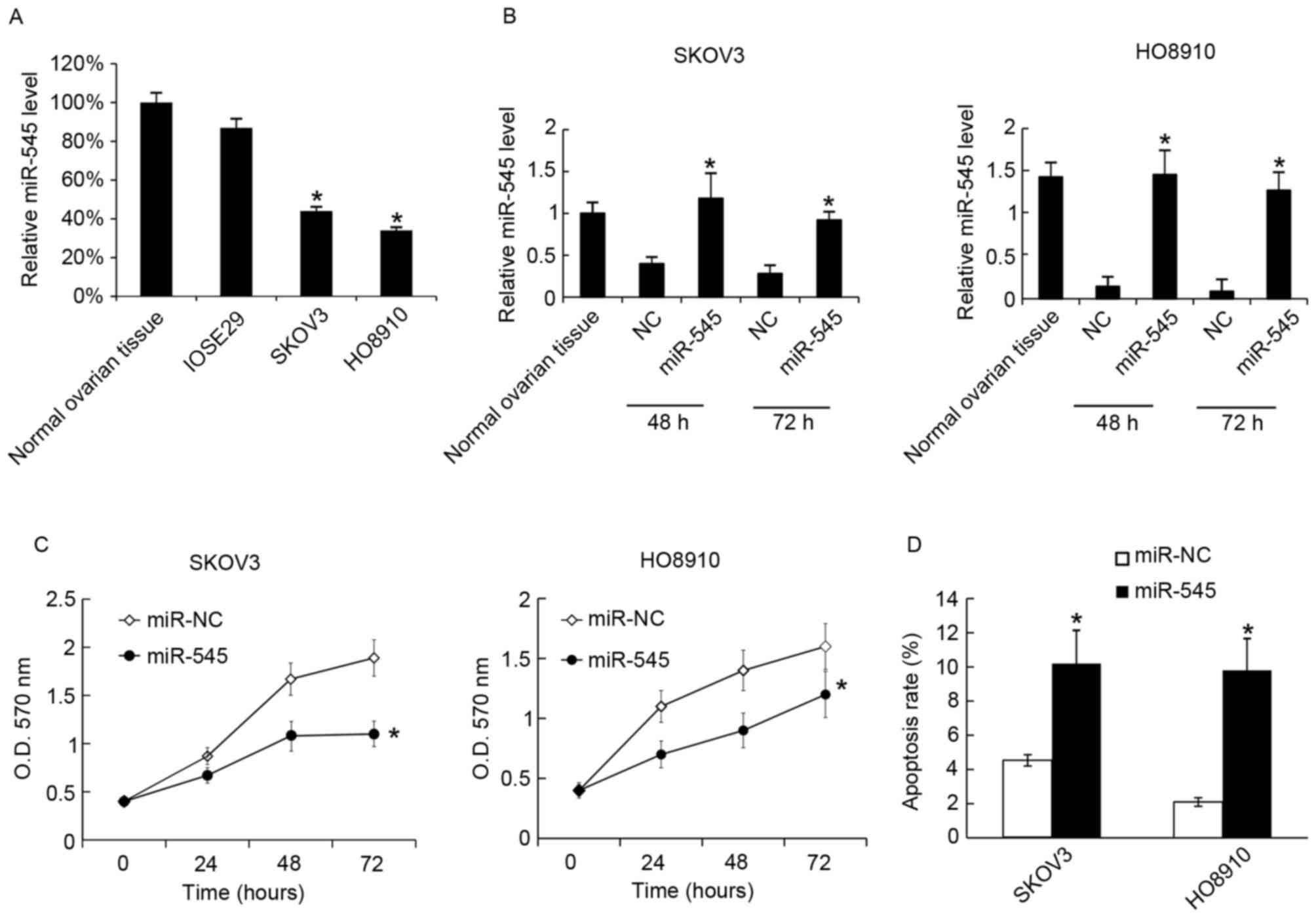 | Figure 2.Transfection of miR-545 mimics
inhibits cell growth and promotes apoptosis. (A) The miR-545 levels
in normal ovarian tissues; IOSE29, SKOV3 and HO8910 cells were
analyzed using RT-qPCR. The miR-545 level in normal ovarian tissues
was arbitrarily defined as 100%. (B) miR-545 mimics were
transfected into SKOV3 and HO8910, and after 48 and 72 h the
miR-545 levels were analyzed using RT-qPCR. Levels of miR-545
expression in normal ovarian tissues was arbitrarily defined as
100%. (C) Following transfection with miR-NC or miR-545 mimic,
cellular proliferation was analyzed using an MTT assay at 24, 48
and 72 h, (D) At 48 h after transfection with the miR-545 mimic,
cell apoptosis was measured using annexin V-propidium iodide
staining. Data are presented as the mean ± standard deviation. The
experiment was repeated at least three times. *P<0.05 vs. NC.
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; miR-545, microRNA-545; OD, optical density; NC, negative
control. |
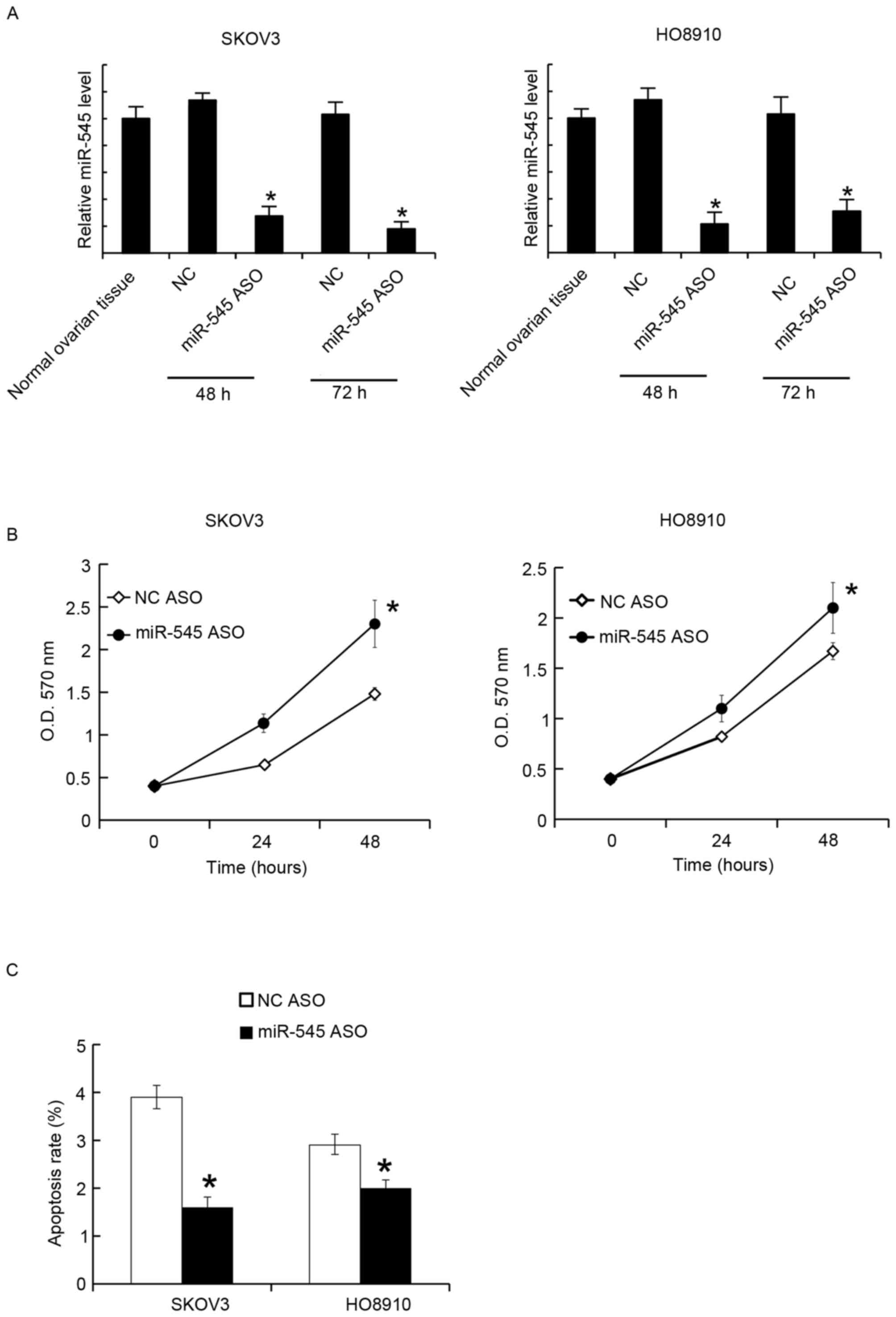
Suppression of miR-545 promoted cell
proliferation and inhibited apoptosis
The miR-545 levels in SKOV3 and HO8910 were
suppressed by miR-545 ASO transfection. After 48 and 72 h, the
miR-545 levels were assayed using RT-qPCR. It was identified that
miR-545 ASO significantly suppressed the miR-545 levels at the two
time points (Fig. 3A). The MTT assay
revealed that miR-545 ASO transfection promoted cellular
proliferation in SKOV3 and HO8910 cells (Fig. 3B). Similarly, 48 h after transfection,
the cell apoptosis rates were assessed using FACS analysis, and it
was revealed that miR-545 ASO decreased the apoptosis rate in SKOV3
and HO8910 cells (Fig. 3C).
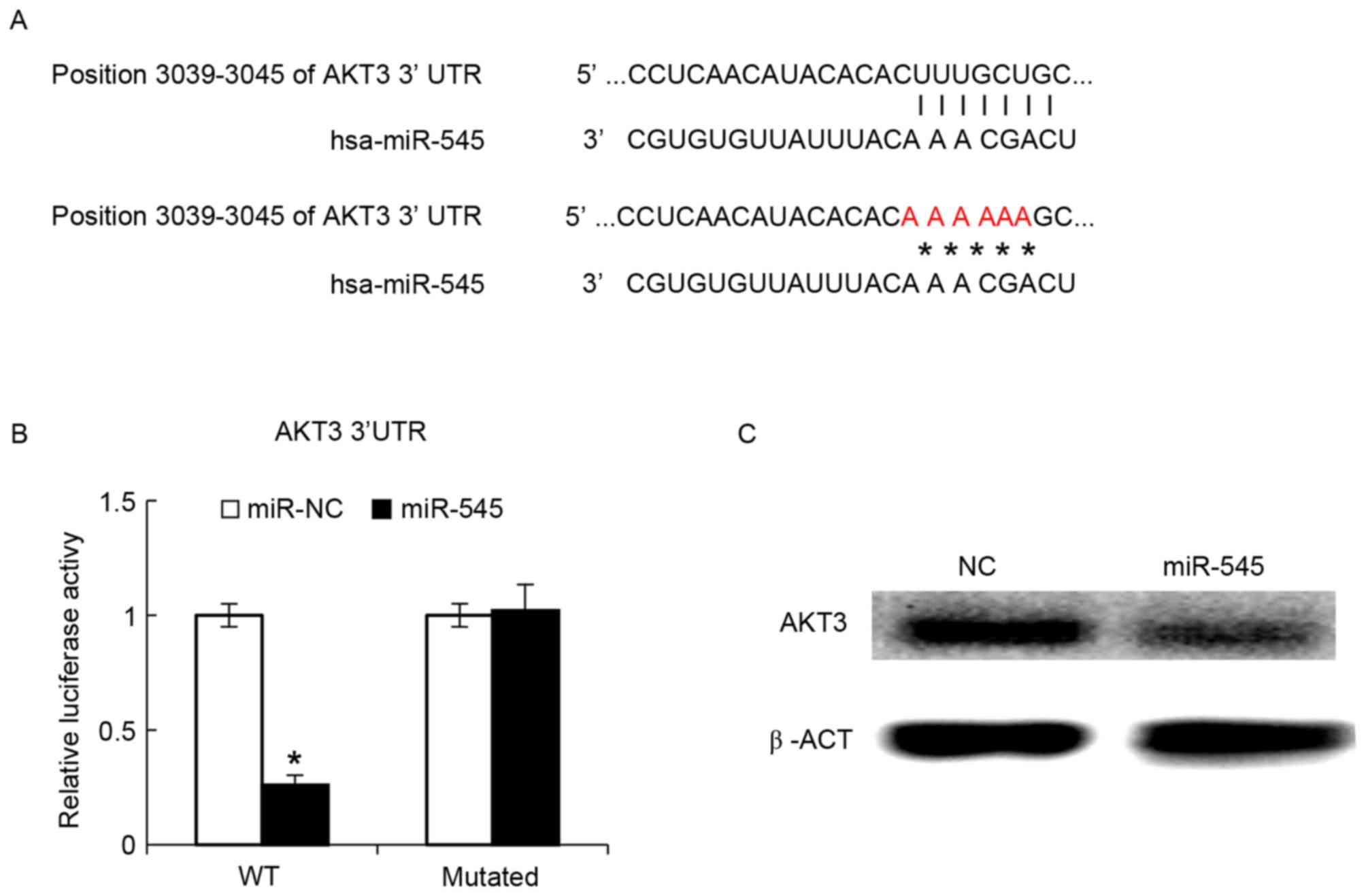
AKT3 is targeted by miR-545
The phosphatidylinositol 3-kinase (PI3K)/AKT pathway
has been demonstrated to serve a function in oncogenic
transformation in ovarian cancer, with AKT3 being an important
mediator of ovarian oncogenesis (29). The bioinformatics algorithm suggested
that miR-545 may bind the 3′-UTR of AKT3 (Fig. 4A). The intact 3′-UTR of AKT3 and its
mutated version were cloned into luciferase reporter plasmids and
used for co-transfection with miR-545 into IOSE29 cells. It was
revealed that miR-545 decreased the luciferase activity of the wild
type 3′-UTR reporter, whereas the luciferase activity of the
reporter with the mutated version was not significantly different
between miR-NC and miR-545 (Fig. 4B).
At 48 h after transfection of the miR-545 mimics into the SKOV3
cells, AKT3 protein levels in the SKOV3 cells were assayed by
western blot analysis, and found that these data indicated that
miR-545 exerted its function through AKT3. Transfected miR-545
mimics decreased the AKT3 protein levels (Fig. 4C).
Discussion
miRNAs have been demonstrated to be involved in the
pathogenesis of EOC (30–32). A previous study by the authors
revealed the function of miR-494 in EOC (33). The authors demonstrated that miR-494
was expressed at low levels in EOC tissues and cells.
Overexpression of miR-494 inhibited the proliferation and migration
of the EOC cells. miR-494 was revealed to target Myc proto-oncogene
protein (34). In the present study,
the function of miR-545 in EOC was analyzed and it was demonstrated
that miR-545 served an antitumor function, and AKT3 was targeted by
miR-545.
miR-545 appears to function as a cancer suppressor
gene in a number of types of cancer, including pancreatic ductal
adenocarcinoma and lung cancer (22,35). In a
previous study, inhibition of miR-545 expression decreased the
radiosensitivity of Lewis lung carcinoma, with miR-545 regulating
Ku70 expression by targeting the 3′-UTR of Ku70, a process that was
involved in radiotherapy (36). On
the one hand, miR-545 can inhibit the proliferation of cancer cells
by targeting oncogenes (35);
however, miR-545 could also enhance radiosensitivity during cancer
therapy. Thus, miR-545 is a good candidate target for cancer
therapy.
Expression of AKT3 is increased in several types of
cancer (37). In triple negative
breast cancer (TNBC), the downregulation of AKT3 inhibits the
cellular proliferation of TNBC cell lines and depletion of AKT3 in
TNBC sensitizes cells to the pan-AKT inhibitor GSK690693 (38). Additionally, in prostate cancer, AKT3
promotes the proliferation of prostate cancer proliferation cells
through regulation of AKT, serine/threonine protein kinase B-Raf
and tuberous sclerosis 1/2 (39).
In conclusion, the data collected in the present
study demonstrated that miR-545 may serve an antitumor function in
EOC.
Acknowledgements
The authors would like to thank Ms Gao Hui (Key
Laboratory of Birth Defects and Related Diseases of Women and
Children, Ministry of Education, West China Second University
Hospital, Sichuan University, Chengdu, China) for her technical
assistance.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Author's contributions
XJ collected patient data and performed cell
experiments. XL analyzed the data. ML performed the statistical
analysis. YZ performed the literature search and analyzed the data.
ZF performed cell experiments. XS performed flow cytometry. YH
collected and analyzed the data. MC contributed to study design and
submitted the manuscript. XY contributed to study design and
manuscript writing.
Ethics approval and consent to
participate
The present study was approved by Ethic Committee of
Sichuan University and each patient provided written informed
consent.
Consent for publication
All samples were collected with the informed consent
of the patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Copeland LJ: Epithelial ovarian cancer.
Clin Gynecol Oncol. 7:313–367. 2007. View Article : Google Scholar
|
|
2
|
Holschneider CH and Berek JS: Ovarian
cancer: Epidemiology, biology, and prognostic factors. Semin Surg
Oncol. 19:3–10. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang B, Liu SZ, Zheng RS, Zhang F, Chen WQ
and Sun XB: Time trends of ovarian cancer incidence in China. Asian
Pac J Cancer Prev. 15:191–193. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shushan A, Paltiel O, Iscovich J, Elchalal
U, Peretz T and Schenker JG: Human menopausal gonadotropin and the
risk of epithelial ovarian cancer. Fertil Steril. 65:13–18. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Daniilidis A and Karagiannis V: Epithelial
ovarian cancer. Risk factors, screening and the role of
prophylactic oophorectomy. Hippokratia. 11:63–66. 2007.PubMed/NCBI
|
|
6
|
Survival of Cancer Patients in Europe: The
EUROCARE-2 study. IARC Sci Publ 1–572. 1999.
|
|
7
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in Cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong
Q, Qin L, Wu X, Zheng Y, Yang Y, et al: Identification of miRNomes
in human liver and hepatocellular carcinoma reveals miR-199a/b-3p
as therapeutic target for hepatocellular carcinoma. Cancer Cell.
19:232–243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li D, Liu X, Lin L, Hou J, Li N, Wang C,
Wang P, Zhang Q, Zhang P, Zhou W, et al: MicroRNA-99a inhibits
hepatocellular carcinoma growth and correlates with prognosis of
patients with hepatocellular carcinoma. J Biol Chem.
286:36677–36685. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mendell JT and Olson EN: MicroRNAs in
stress signaling and human disease. Cell. 148:1172–1187. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Resnick KE, Alder H, Hagan JP, Richardson
DL, Croce CM and Cohn DE: The detection of differentially expressed
microRNAs from the serum of ovarian cancer patients using a novel
real-time PCR platform. Gynecol Oncol. 112:55–59. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chung YW, Bae HS, Song JY, Lee JK, Lee NW,
Kim T and Lee KW: Detection of microRNA as novel biomarkers of
epithelial ovarian cancer from the serum of ovarian cancer
patients. Int J Gynecol Cancer. 23:673–679. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shapira I, Oswald M, Lovecchio J, Khalili
H, Menzin A, Whyte J, Dos Santos L, Liang S, Bhuiya T, Keogh M, et
al: Circulating biomarkers for detection of ovarian cancer and
predicting cancer outcomes. Br J Cancer. 110:976–983. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Suryawanshi S, Vlad AM, Lin HM,
Mantia-Smaldone G, Laskey R, Lee M, Lin Y, Donnellan N, Klein-Patel
M, Lee T, et al: Plasma microRNAs as novel biomarkers for
endometriosis and endometriosis-associated ovarian cancer. Clin
Cancer Res. 19:1213–1224. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng H, Zhang L, Zhao Y, Yang D, Song F,
Wen Y, Hao Q, Hu Z, Zhang W and Chen K: Plasma miRNAs as diagnostic
and prognostic biomarkers for ovarian cancer. PLoS One.
8:e778532013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kan CW, Hahn MA, Gard GB, Maidens J, Huh
JY, Marsh DJ and Howell VM: Elevated levels of circulating
microRNA-200 family members correlate with serous epithelial
ovarian cancer. BMC Cancer. 12:6272012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Giordano S and Columbano A: MicroRNAs: New
tools for diagnosis, prognosis, and therapy in hepatocellular
carcinoma? Hepatology. 57:840–847. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song B, Ji W, Guo S, Liu A, Jing W, Shao
C, Li G and Jin G: miR-545 inhibited pancreatic ductal
adenocarcinoma growth by targeting RIG-I. FEBS Lett. 588:4375–4381.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Du B, Wang Z, Zhang X, Feng S, Wang G, He
J and Zhang B: MicroRNA-545 suppresses cell proliferation by
targeting cyclin D1 and CDK4 in lung cancer cells. PLoS One.
9:e880222014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yiwei T, Hua H, Hui G, Mao M and Xiang L:
HOTAIR interacting with MAPK1 regulates ovarian cancer skov3 cell
proliferation, migration, and invasion. Med Sci Monit.
21:1856–1863. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song B, Zhang C, Li G, Jin G and Liu C:
MiR-940 inhibited pancreatic ductal adenocarcinoma growth by
targeting MyD88. Cell Physiol Biochem. 35:1167–1177. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou Q and Yu Y: Upregulated CDK16
expression in serous epithelial ovarian cancer cells. Med Sci
Monit. 21:3409–3414. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li H, Xu Y, Qiu W, Zhao D and Zhang Y:
Tissue miR-193b as a novel biomarker for patients with ovarian
cancer. Med Sci Monit. 21:3929–3934. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife: Aug. 12:42015.
|
|
29
|
Cristiano BE, Chan JC, Hannan KM, Lundie
NA, Marmy-Conus NJ, Campbell IG, Phillips WA, Robbie M, Hannan RD
and Pearson RB: A specific role for AKT3 in the genesis of ovarian
cancer through modulation of G2-M phase transition. Cancer
Research. 66:11718–11725. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang L, Volinia S, Bonome T, Calin GA,
Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K,
et al: Genomic and epigenetic alterations deregulate microRNA
expression in human epithelial ovarian cancer. Proc Natl Acad Sci
USA. 105:pp. 7004–7009. 2008; View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang H, Kong W, He L, Zhao JJ, O'Donnell
JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV and Cheng JQ:
MicroRNA expression profiling in human ovarian cancer: miR-214
induces cell survival and cisplatin resistance by targeting PTEN.
Cancer Res. 68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li N, Zhao X, Wang L, Zhang S, Cui M and
He J: miR-494 suppresses tumor growth of epithelial ovarian
carcinoma by targeting IGF1R. Tumor Biol. 37:7767–7776. 2016.
View Article : Google Scholar
|
|
34
|
Yuan J, Wang K and Xi M: MiR-494 inhibits
epithelial ovarian cancer growth by targeting c-Myc. Med Sci Monit.
22:617–624. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Song B, Ji W, Guo S, Liu A, Jing W, Shao
C, Li G and Jin G: miR-545 inhibited pancreatic ductal
adenocarcinoma growth by targeting RIG-I. FEBS Lett. 588:4375–4381.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liao C, Xiao W, Zhu N, Liu Z, Yang J, Wang
Y and Hong M: MicroR-545 enhanced radiosensitivity via suppressing
Ku70 expression in Lewis lung carcinoma xenograft model. Cancer
Cell Int. 15:562015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chin YR, Yoshida T, Marusyk A, Beck AH,
Polyak K and Toker A: Targeting Akt3 signaling in triple-negative
breast cancer. Cancer Res. 74:964–973. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lin HP, Lin CY, Huo C, Jan YJ, Tseng JC,
Jiang SS, Kuo YY, Chen SC, Wang CT, Chan TM, et al: AKT3 promotes
prostate cancer proliferation cells through regulation of Akt,
B-Raf, and TSC1/TSC2. Oncotarget. 6:27097–27112. 2015. View Article : Google Scholar : PubMed/NCBI
|