Introduction
Gastric cancer is one of the most common types of
adenocarcinoma and the second leading cause of cancer-associated
cases of mortality worldwide, despite having a low incidence in
developed countries (1). The therapy
for stomach cancer is an integrated process that involves surgery,
radiation treatment and chemotherapy (2). It has been acknowledged that the
effectiveness of treatment of tumors is associated with TNM-staging
(3); however, patients with gastric
carcinoma may be treated with radical resection, but the 5-year
survival rates of patients are low due to the invasiveness and drug
resistance of cancer cells (4). Thus,
there is an urgent requirement to study the molecular mechanisms of
gastric carcinoma (5). MicroRNAs
(miRNAs) are small, non-coding RNAs composed of 18–25 nucleotides,
which function through binding the 3′-untranslated region (3′-UTR)
of targeted mRNAs, eventually leading to decreased gene expression
and translation (6). It has been
reported that abnormal levels of miRNAs are associated with the
occurrence of cancer via affecting cell proliferation, apoptosis,
metastasis and susceptibility to therapy (7). Therefore, studying the function of
miRNAs may reveal novel treatment strategies for cancer.
miRNA (miR)-130b, situated in the 22q11 locus, has
been demonstrated to have dual characteristics in the progression
of tumors. For instance, it was identified to be an oncogene in
gastric cancer, liver cancer, and endometrial cancer. By contrast,
it was demonstrated to have a protective effect against ovarian and
thyroid papillary carcinoma (8).
These studies indicate that the role of miR-130b depends on the
tumor classification. Our previous study identified that the
expression of RAS protein activator like 1 (RASAL1) is decreased in
gastric cancer in in vitro and in vivo experiments
(9). However, the association between
miR-130b-5p and RASAL1 and the effect of miR-130b-5p on gastric
cancer has not previously been studied.
In the present study, an increase in miR-130b-5p and
a decrease in RASAL1 were identified in gastric carcinoma cell
lines. In addition, miR-130b-5p could facilitate the migration and
invasion abilities of gastric cell lines. The role of miR-130b-5p
in gastric cancer was associated with decreased RASAL1, which was
demonstrated to be a novel target gene of miR-130b-5p. These
findings could have implications for improving the treatment of
gastric cancer.
Materials and methods
Cell lines and culture
The human gastric cell lines AGS, BGC823, MGC803,
MKN45, MKN74, SGC7901 and GES-1 were purchased from Shanghai
Institute of Digestive Disease (Shanghai, China) and were cultured
in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), supplemented with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.), 100 µg/ml streptomycin and 100
U/ml penicillin (Gibco; Thermo Fisher Scientific, Inc.). Cells were
cultured in a 37°C incubator with 5% CO2.
Transfection of MGC803 cells with
miR-130b-5p mimics and inhibitor
MGC803 cells were transfected with miR-130b-5p
mimics (sense, 5′-ACUCUUUCCCUGUUGCACUAC-3′ and antisense,
5′-AGUGCAACAGGGAAAGAGUUU-3′) and miR-130b-5p inhibitor
(GUAGUGCAACAGGGAAAGAGU). Cells were transfected with NC
(5′-UCACAACCUCCUAGAAAGAGU-3′) as the control. miR-130b-5p mimics,
miR-130b-5p inhibitor and NC were designed and synthesized by
Shanghai GenePharma Co., Ltd. (Shanghai, China) and transfected
into cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Transfection was conducted for 1–2 days according to cell growth
conditions prior to subsequent analysis. Each experiment was
repeated at least three times.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Expression of miR-130b-5p and RASAL1 were detected
by RT-qPCR. TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) was
used to extract total RNA from cell cultures, with GES-1 as the
control, then cDNA was obtained by RT using PrimeScript cDNA qPCR
RT kit (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Then, the acquired cDNA was used as a
template to amplify double-stranded DNA using FastStart Universal
SYBR-Green Master mix (Shanghai GenePharma, Co., Ltd.). Primers
used for qPCR were as follows: β-actin (upstream,
5′-CTACAATGAGCTGCGTGTGG-3′ and downstream,
5′-TAGCTCTTCTCCAGGGAGGA-3′, 221 bp), RASAL1 (upstream,
5′-TGGATTTCTCTTCTTGCGATTCT-3′ and downstream,
5′-TGTTGGTCCCGAAGGTCAA-3′, 72 bp), miR-130b (upstream,
5′-GCCGCCAGTGCAATGATGAA-3′ and downstream, 5′-GTGCAGGGTCCGAGGT-3′);
and U6 (upstream, 5′-CGCTTCGGCAGCACATATACTA-3′ and downstream,
5′-CGCTTCACGAATTTGCGTGTCA-3′). The reaction conditions were as
follows: 95°C for 2 min, 40 cycles at 95°C for 30 sec, 58°C for 30
sec and 72°C for 1 min, using an ABI Step One PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The expression of U6
was used as internal control. The relative expression of
miR-130b-5p and RASAL1 compared with U6 was calculated using the
2−ΔΔCq method (10). The
experiment was repeated three times.
Western blotting detection of
RASAL1
To demonstrate the effect of miR-130b-5p on RASAL1
protein expression, MGC803 cells were transfected with miR-130b-5p
mimics or miR-130b-5p inhibitor, with cells transfected with
negative control (NC) sequence as the control. The transfected
cells were collected, dissociated and incubated with
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Shanghai, China) for protein extraction. Protein
concentration was measured by the bicinchoninic acid (BCA) method.
The expression of RASAL1 protein was measured by western blot
analysis. Protein supernatants were separated via 10% SDS-PAGE
(Invitrogen; Thermo Fisher Scientific, Inc.) and transferred to
nitrocellulose membranes (placed in ice water with a constant
current of 100 mA overnight). The antibody was blocked with 5% skim
milk at 4°C overnight. Rabbit anti-human RASAL1 polyclonal antibody
(ab214321, 1:1,000; Abcam, Cambridge, UK) was used as the primary
antibody and goat anti-rabbit-IgG (H+L) antibody conjugated with
horseradish peroxidase (ab205718, 1:5,000; Abcam) was used as the
secondary antibody (and incubated for one hour at room
temperature). β-actin (ab8226; Abcam) served as the internal
control. The enhanced chemiluminescence imaging method was used to
facilitate the detection of protein bands. The experiment was
repeated three times.
Luciferase reporter gene assay
The 293T cells (Ribobio Co., Ltd., Guangzhou, China)
were cultured in an incubator at 37°C and 5% CO2. The
pGL3-promoter vector containing the RASAL1 3′UTR and the control
plasmid psiC-2 was constructed by Guangzhou Ribobio Co., Ltd., and
then transfected with miR-130b-5p-mimics and NC, with NC as
control. Then, logarithmic phase cells were plated onto 96-well
plates at a density of 1.5×104 cells/well and incubated
at 37°C for 24 h. Then, 293T cells were transfected with pGL3
promoter RASAL1 and miR-130b-5p-mimics and NC seperately, using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At
6 h, complete medium was injected into the cell culture media and
incubated at 37°C for 48 h to achieve high transfection efficiency.
Then, cells were collected by the trypsin digestion method. A
mixture of 100 µl cell lysate and 100 µl luciferase detection
reagent (Beyotime Institute of Biotechnology) per sample was used
to measure luciferase activity in relative light units, using a
GLOMAX 20/20 luminometer (Promega Corporation, Madison, WI, USA),
according to the manufacturer's protocol. The cell lysis solution
was used as a blank control. The fluorescence value of hRluc
(Hren-luciferase gene) was compared with that of hLuc (Firefly
luciferase gene); the fluorescence ratio of the experimental group
and the control group were analyzed. Each experiment was repeated
at least three times.
Colony formation assay
Cells transfected with miR-130b-5p mimics, inhibitor
and NC (control group) were used to evaluate the effect of abnormal
miR-130b-5p expression on proliferation ability in gastric cancer
cells. Cells from each group were seeded into 6-well plates (1,000
cells/well) and incubated for 1 week in RPMI-1640 medium with 10%
FBS, 100 µg/ml streptomycin, and 100 U/ml penicillin. Cells from
each group were stained with crystal violet for 30 min at room
temperature. Clones >2 mm were counted, and the result was
calculated as the mean of three wells. Each experiment was repeated
at least three times.
Cell proliferation assay
Cells transfected with miR-130b-5p mimics,
miR-130b-5p inhibitor and NC (control group) were seeded in a
96--well plate at a density of 2×103 cells/well, and
incubated for 24 h at 37°C for adherence. MTT solution (20 µl in
200 µl media) was added and incubated for 4 h at 37°C. This was
followed by removal of the remaining liquid and injection of 150 µl
DMSO per well. The formazan crystals were dissolved by gentle
shaking of the plate for 5 min at room temperature. The absorbance
was measured at 540 nm using a multi-scan plate reader. Each
experiment was repeated at least three times.
Transwell assays
MGC803 cells transfected with miR-130b-5p mimics and
inhibitor were incubated at 37°C and 5% CO2, with cells
transfected with NC as the control. Matrigel (BD Biosciences,
Franklin Lakes, NJ, USA) was combined with serum-free RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.). For the invasion
assay, the Transwell chamber contained Matrigel. For the migration
assay, no Matrigel was used. Cells from each group were
trypsinized, centrifuged (1,000 rpm, 167.7 × g for 5 min at room
temperature) and resuspended in serum-free medium at a
concentration of 5×105/ml. Serum-free medium (50 µl) was
added to the upper well of the Transwell chamber containing
2.5×103 cells/per well (Corning, Inc., Corning, NY, USA)
with RPMI-1640 medium (containing 10% serum) in the lower site.
Following incubation for 48 h, cells in the upper chamber were
removed and cells in the lower chamber were stained with crystal
violet at room temperature for 30 min. Finally, cells in the lower
chamber were counted under a light microscope. Each experiment was
repeated at least three times.
Statistical analysis
All experimental data were analyzed using SPSS 18.0
software (SPSS, Inc., Chicago, IL, USA) and presented as the mean ±
standard deviation. Comparisons between experimental and control
groups were performed using Student's t-tests. Significant
differences among groups were assessed by one-way analysis of
variance followed by Tukey's post hoc test for multiple
comparisons. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression of miR-130b-5p and RASAL1
in seven gastric cell lines
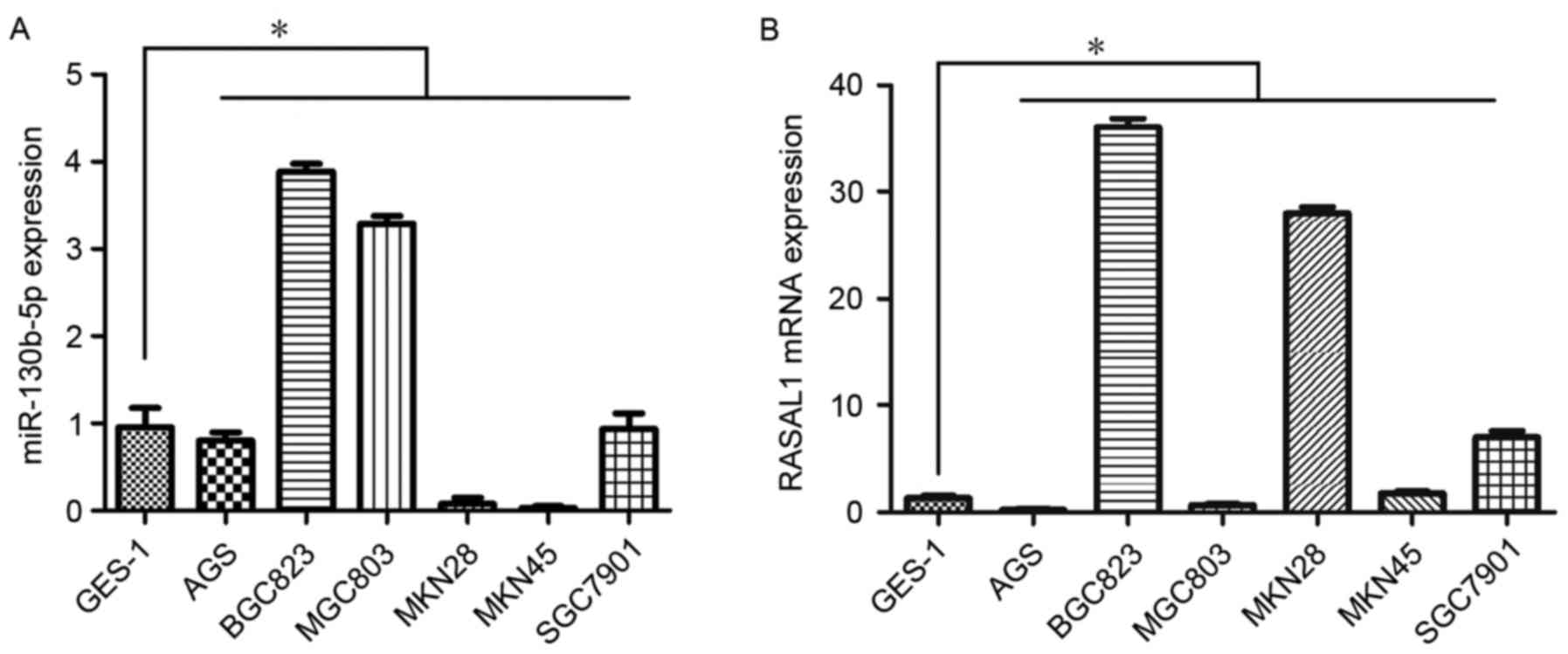
Expression of miR-130b-5p and RASAL1 was assessed by
RT-qPCR in seven gastric cell lines. The results demonstrated
higher expression of miR-130b-5p in BGC823 cells and MGC803 cells
compared with the rest of the gastric cell lines studied (Fig. 1A). Furthermore, consistent with the
higher expression of miR-130b-5p, the expression of RASAL1 was
markedly decreased in MGC803 cells compared with other cell lines
(Fig. 1B). As a result, the MGC803
cell line was selected for subsequent experiments.
RASAL1 is a target gene of
miR-130b-5p
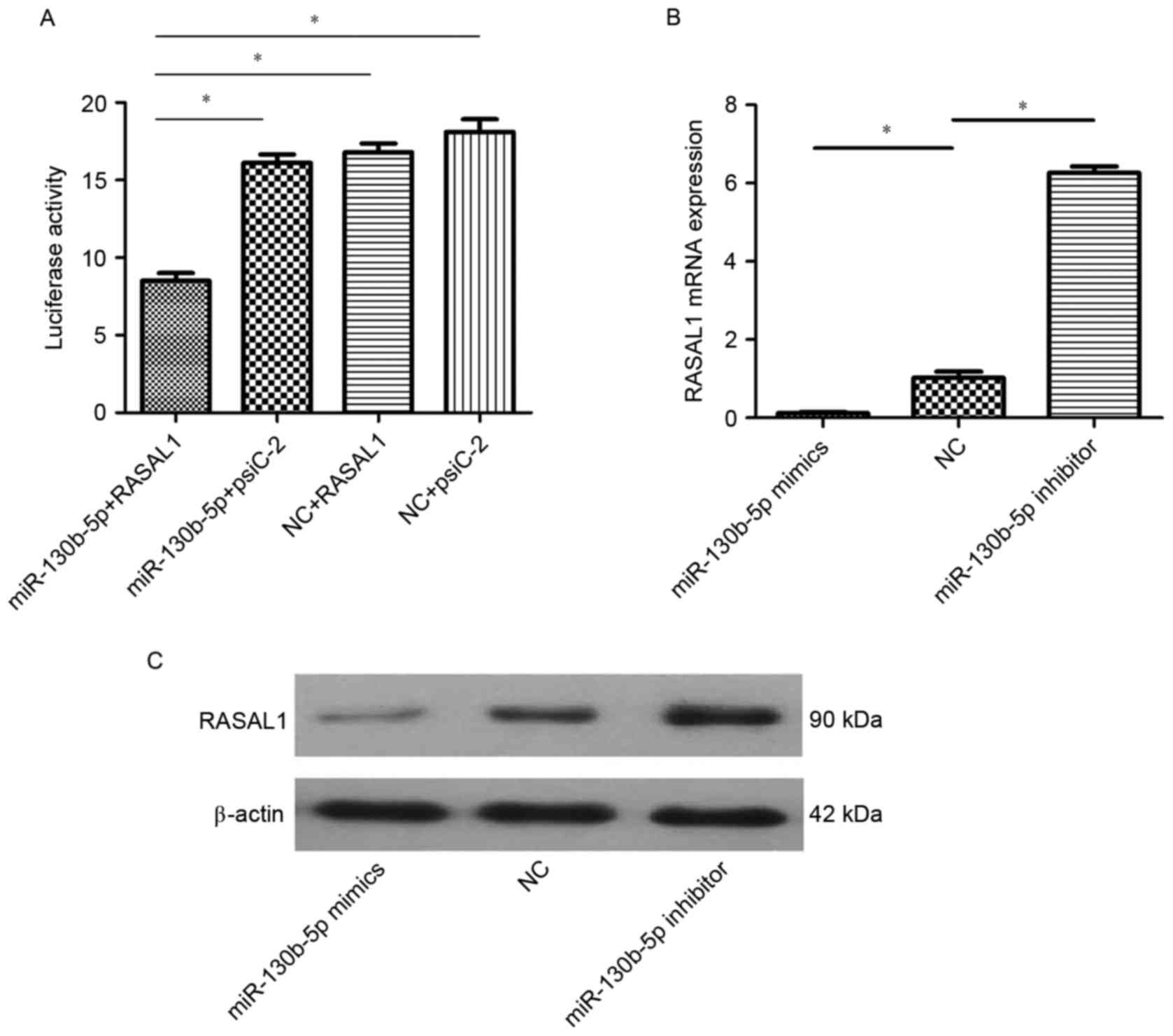
In order to reveal the molecular mechanism of
miR-130b-5p in gastric cancer, bioinformatics software (FindTar3,
version 3.11.12; https://bio.sz.tsinghua.edu.cn) was used to predict
the targeted interaction between RASAL1 and miR-130b-5p. The 8–29
and 684–707 loci of the 3′-UTR of RASAL1 were predicted to be a
complementary target of miR-130b-5p. Based on this prediction, a
luciferase reporter gene assay was used to evaluate the interaction
between RASAL1 and miR-130b-5p. A significant decrease in
luciferase activity was observed in cells co-transfected with
miR-130b-5p and pGL3-RASAL1 compared with the other three groups
(P<0.05; Fig. 2A). These results
demonstrated that RASAL1 may be regulated by miR-130b-5p. To verify
these findings, the mRNA and protein expression of RASAL1 was
measured in cells transfected with miR-130b-5p mimics, miR-130b-5p
inhibitor and NC by RT-qPCR and western blot analysis. The results
indicated decreased expression of RASAL1 at the mRNA protein levels
in cells transfected with miR-130b-5p mimics (P<0.05) and a
significant increase in RASAL1 in cells transfected with
miR-130b-5p inhibitor (P<0.05), compared with cells transfected
with NC (Fig. 2B and C).
Cell proliferation assay
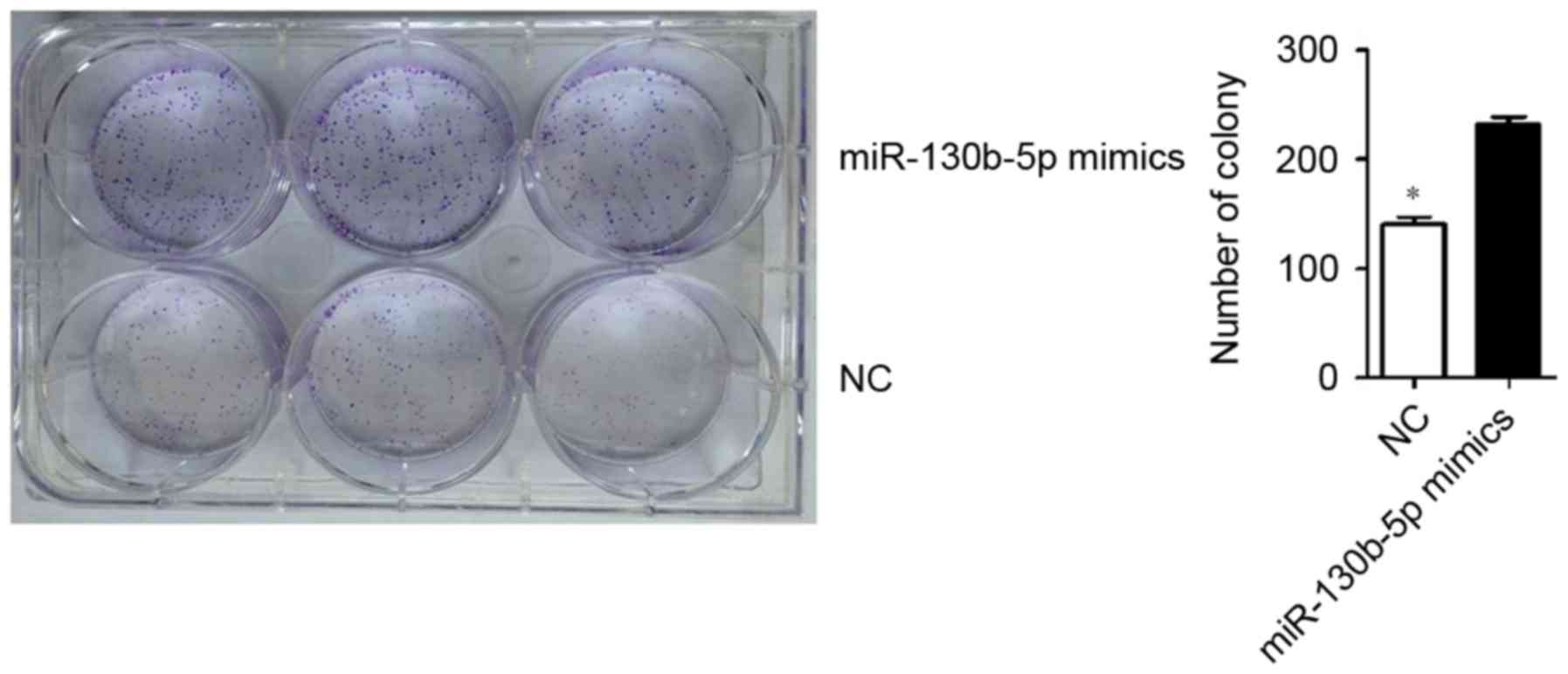
MTT assay and low-density colony formation
experiments were used to evaluate the effect of miR-130b-5p on
MGC803 cell proliferation. The results revealed a significantly
higher clone formation ability in cells transfected with
miR-130b-5p mimics (P<0.05; Fig.
3) and a significantly lower clone formation ability in cells
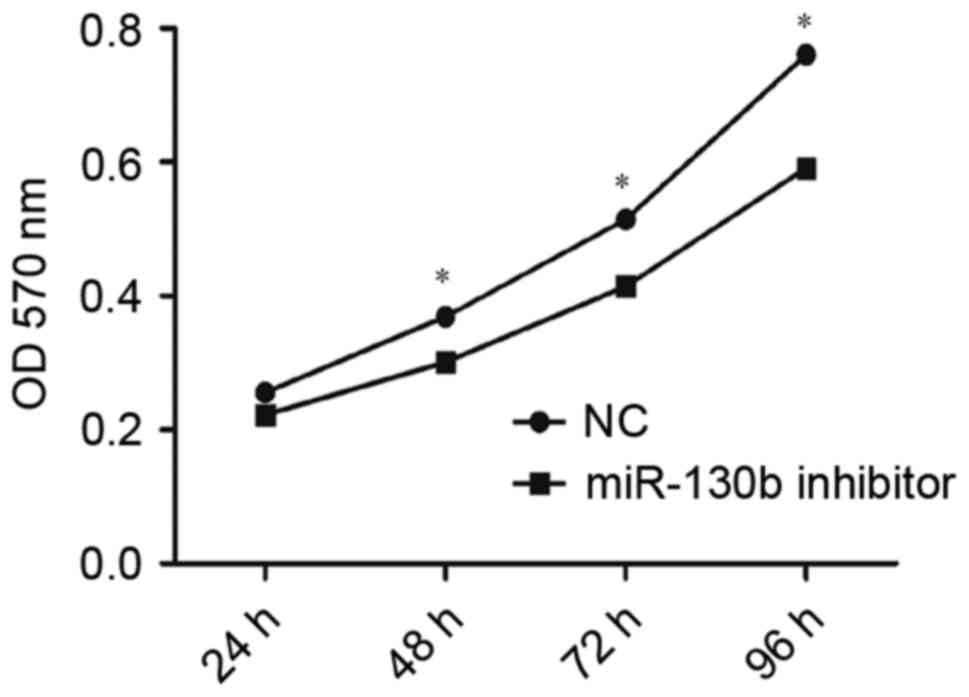
transfected with miR-130b-5p inhibitor (P<0.05; Fig. 4) compared with the NC group.
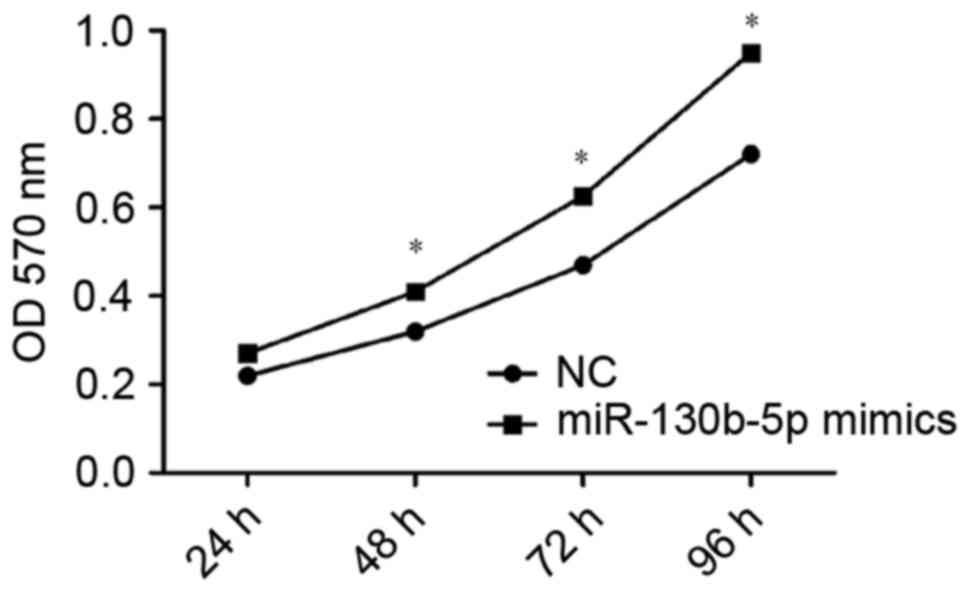
Consistent with this, OD values indicated a significantly higher
rate of proliferation in cells transfected with miR-130b-5p mimics
(P<0.05; Fig. 5) and a
significantly lower rate of proliferation in cells transfected with
miR-130b-5p inhibitor (P<0.05; Fig.
6) compared with the NC group at 48, 72 and 96 h. These results
indicated an increased capacity for cell proliferation following
overexpression of miR-130b-5p and a weakened capacity following
downregulation of miR-130b-5p.
Measurement of migration and
invasion
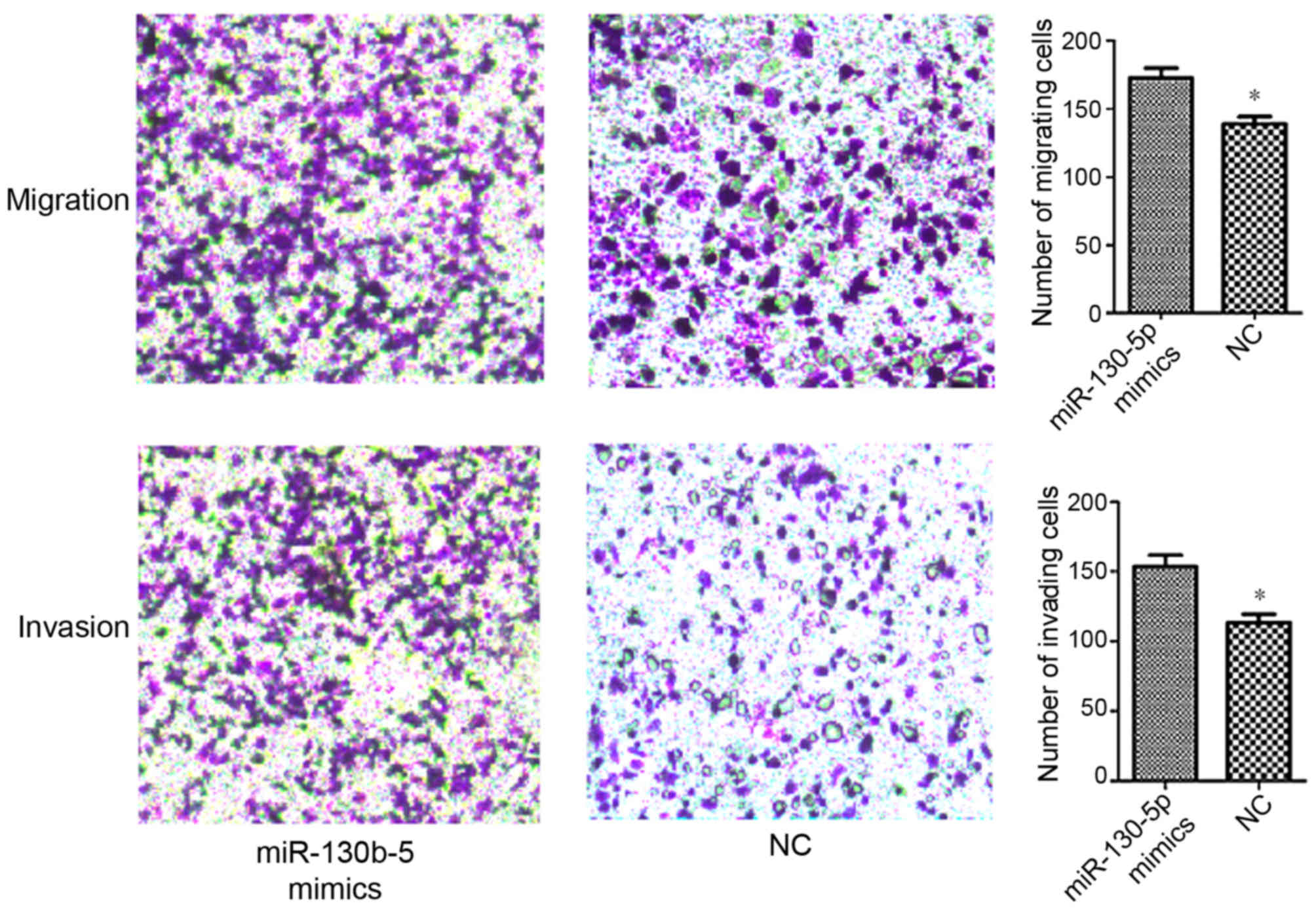
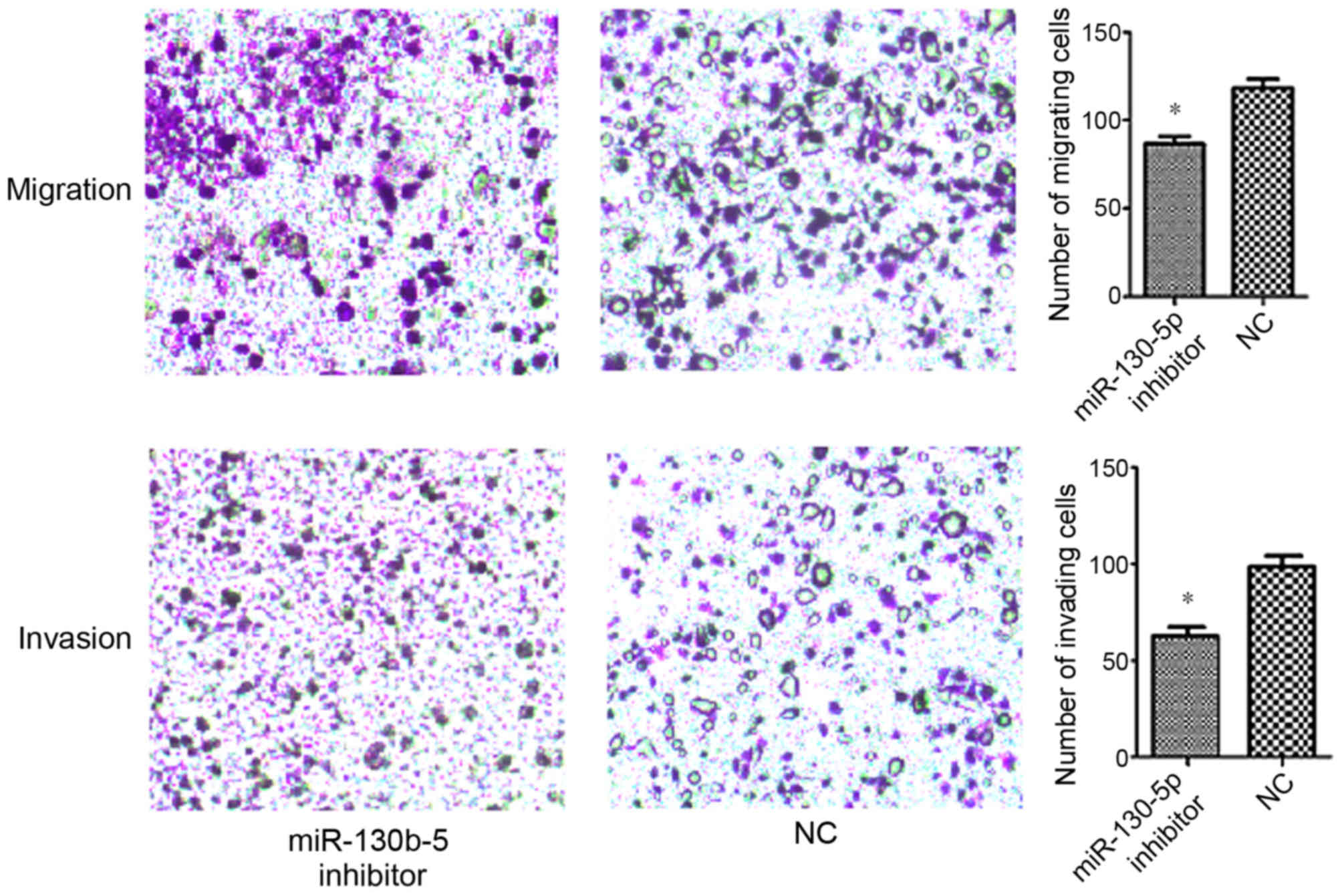
Transwell assays were performed in order to evaluate
the effect of miR-130b-5p on the migration and invasion abilities
of MGC803 cells. The data demonstrated a significantly increased
migration and invasion ability in cells transfected with
miR-130b-5p mimics (P<0.05; Fig.
7), and a significantly decreased migration and invasion
ability in cells transfected with miR-130b-5p inhibitor (P<0.05;
Fig. 8) compared with cells
transfected with NC.
Discussion
The association between RAS gene and occurrence of
cancer was gradually taken seriously in recent studies. The
RAS-RAF-MEK-ERK signal transduction pathway performs a key function
in cell proliferation, migration and invasion (11). In the process of regulation of cell
proliferation, migration and invasion, the product of the activated
RAS gene (RAS protein) is a key controlling element in the
RAS-RAF-MEK-ERK signal transduction pathway. It has been reported
that RAS gene mutations eventually lead to abnormal RAS
hyperactivity, excessive cell growth and accelerated development of
tumors (12).
RAS proteins (Ras, Rho, Rab, Arf, Sar1 and Ran
families) are an element of GTP hydrolases (G proteins) that
regulate and control the RAS hyperactivity. The RAS protein exists
in two forms; an active form bound with GTP and an inactive form
bound with GDP. The transformation of RAS protein structures is
closely associated with the capacity for cell proliferation,
migration and invasion in gastric cell lines (5). There are two types of molecule involved
in the conversion between the active and inactive forms of RAS
protein: RAS GAPs and RAS GEFs. The function of RAS GEFs is to
facilitate binding with GTP and eventually lead to the activation
of RAS, whereas RAS GAPs promote the conversion from GTP binding to
GDP binding and ultimately give rise to inactivation of RAS
(13). RASAL1 is a key member of the
RAS GAP family (14). RASAL1 is
thought to act as a tumor suppressor gene by promoting the
conversion from GTP binding (active) to GDP binding (inactive),
eventually suppressing tumorigenesis in multiple types of cancer
(15). A previous study indicated
that various types of cancer are associated with low expression of
the RASAL1 gene, including nasopharyngeal carcinoma, breast cancer,
lung cancer, liver cancer and esophageal cancer (13).
Our previous research indicated that overexpression
of RASAL1 may attenuate gastric carcinogenesis in vivo and
in vitro (9). In other words,
RASAL1 is a tumor suppressor gene and low expression of RASAL1 may
result in RAS hyperactivity, leading to the occurrence of malignant
tumors. miR-130b-5p is a small non-coding RNA situated in the 22q11
locus. Numerous studies have indicated that miR-130b is
overexpressed in various types of tumor, including gastric cancer,
liver cancer and endometrial cancer (16–18). It
was verified that the oncogenic role of miR-130b-5p is achieved by
means of downregulation of a diverse set of cancer suppressor
genes, including phosphatase and tensin homolog serve protective
roles against esophageal cancer (19). In the present study, bioinformatics
software (FindTar3) was used to predict RASAL1 as an miR-130b-5p
target gene and a luciferase reporter gene assay was used to verify
the targeted interaction between miR-130b-5p and RASAL1. In
addition, a low level of RASAL1 expression was observed by RT-qPCR
and western blot analysis following transfection with miR-130b-5p
mimics. Conversely, a high level of RASAL1 expression was observed
in cells transfected with miR-130b-5p inhibitor. The results also
demonstrated that the level of miR-130b-5p was higher in MGC803
cells compared with other gastric cell lines. A high level of
miR-130b-5p expression accelerated RAS hyperactivity and suppressed
the expression of RASAL1 gene, eventually resulting in
tumorigenesis. On the other hand, downregulated miR-130b-5p in
cells transfected with miR-130b-5p inhibitor was demonstrated to
exhibit a protective effect by accelerating the expression of
cancer suppressor gene RASAL1. It has been reported that activation
of the RAS-RAF-MEK-ERK transduction pathway is of vital importance
in cell proliferation, migration and invasion (9). The current data demonstrated that a high
level of miR-130b-5p expression could restrain the expression of
RASAL1 and accelerate the proliferation, invasion and migration
ability of MGC803 cells. By contrast, increased RASAL1 and reduced
ability of proliferation, invasion and migration were observed in
cells with downregulated miR-130b-5p.
In conclusion, the present study described the
miR-130b-5p/RASAL1 axis and provided a potential mechanism for RAS
hyperactivity. In addition, the present study identified the effect
of miR-130b-5p in gastric cell proliferation, migration and
invasion. It has been demonstrated that miR-130b-5p is involved in
the occurrence and development of gastric cancer by targeting
RASAL1. These results offer a potential basis for novel targeted
therapy in gastric cancer. The current findings will be explored
further via experimentation on animals in future studies.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Research
Project of National Health and Family Planning Commission of China
(grant no. W201305) and the Natural Science Foundation of Jiangsu
Province of China (grant no. BK2008301).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HC designed the study. YY performed the experiments.
DS and JZ conducted luciferase assays. QY conducted the cell
formation assay. YY and JW analyzed the data.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
Professor Hong Chen, Department of Gastroenterology,
Zhongda Hospital Medical School, Southeast University, 87
Dingjiaqiao, Gulou, Nanjing, Jiangsu 210000, P.R. China
References
|
1
|
Shin JY, Kim YI, Cho SJ, Lee MK, Kook MC,
Lee JH, Lee SS, Ashktorab H, Smoot DT, Ryu KW, et al: MicroRNA 135a
suppresses lymph node metastasis through down-regulation of ROCK1
in early gastric cancer. PLoS One. 9:e852052014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Song Z, Wu Y, Yang J, Yang D and Fang X:
Progress in the treatment of advanced gastric cancer. Tumour Biol.
39:2017. View Article : Google Scholar
|
|
3
|
Wittekind C: The development of the TNM
classification of gastric cancer. Pathol Int. 65:399–403. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim SJ, Wang YG, Lee HW, Kang HG, La SH,
Choi IJ, Irimura T, Ro JY, Bresalier RS and Chun KH: Up-regulation
of neogenin-1 increases cell proliferation and motility in gastric
cancer. Oncotarget. 5:3386–3398. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen H, Cheng ZY, Pan Y, Wang Z, Liu Y and
Zhang JQ: RASAL1 influences the proliferation and invasion of
gastric cancer cells by regulating the RAS/ERK signaling pathway.
Hum Cell. 27:103–110. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gong B, Liu WW, Nie WJ, Li DF, Xie ZJ, Liu
C, Liu YH, Mei P and Li ZJ: MiR-21/RASA1 axis affects malignancy of
colon cancer cells via RAS pathways. World J Gastroenterol.
21:1488–1197. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hao NB, He YF, Li XQ, Wang K and Wang RL:
The role of miRNA and lncRNA in gastric cancer. Oncotarget.
8:81572–81582. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tu K, Zheng X, Dou C, Li C, Yang W, Yao Y
and Liu Q: MicroRNA-130b promotes cell aggressiveness by inhibiting
peroxisome proliferator-activated receptor gamma in human
hepatocellular carcinoma. Int J Mol Sci. 15:20486–20499. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen H, Zhao JY, Qian XC, Cheng ZY, Liu Y
and Wang Z: RASAL1 attenuates gastric carcinogenesis in nude mice
by blocking RAS/ERK signaling. Asian Pac J Cancer Prev.
16:1077–1082. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McCubrey JA, Steelman LS, Chappell WH,
Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M,
Tafuri A, et al: Roles of the Raf/MEK/ERK pathway in cell growth,
malignant transformation and drug resistance. Biochim Biophys Acta.
1773:1263–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen H, Pan Y, Cheng ZY, Wang Z, Liu Y,
Zhao ZJ and Fan H: Hypermethylation and Clinicopathological
significance of RASAL1 gene in gastric cancer. Asian Pac J Cancer
Prev. 14:6261–6265. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jin H, Wang X, Ying J, Wong AH, Cui Y,
Srivastava G, Shen ZY, Li EM, Zhang Q, Jin J, et al: Epigenetic
silencing of a Ca(2+)-regulated Ras GTPase-activating protein RASAL
defines a new mechanism of Ras activation in human cancers. Proc
Natl Acad Sci USA. 104:12353–12358. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Calvisi DF, Ladu S, Conner EA, Seo D,
Hsieh JT, Factor VM and Thorgeirsson SS: Inactivation of Ras
GTPase-activating proteins promotes unrestrained activity of
wild-type Ras in human liver cancer. J Hepatol. 54:311–319. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bernards A and Settleman J: Loss of the
Ras regulator RASAL1: Another route to Ras activation in colorectal
cancer. Gastroenterology. 136:46–48. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lai KW, Koh KX, Loh M, Tada K, Subramaniam
MM, Lim XY, Vaithilingam A, Salto-Tellez M, Iacopetta B, Ito Y, et
al: MicroRNA-130b regulates the tumour suppressor RUNX3 in gastric
cancer. Eur J Cancer. 46:1456–1463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li BL, Lu C, Lu W, Yang TT, Qu J, Hong X
and Wan XP: miR-130b is an EMT-related microRNA that targets DICER1
for aggression in endometrial cancer. Med Oncol. 30:4842013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Colangelo T, Fucci A, Votino C, Sabatino
L, Pancione M, Laudanna C, Binaschi M, Bigioni M, Maggi CA, Parente
D, et al: MicroRNA-130b promotes tumor development and is
associated with poor prognosis in colorectal cancer. Neoplasia.
15:1086–1099. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu T, Cao R, Li S, Fu M, Ren L, Chen W,
Zhu H, Zhan Q and Shi R: MiR-130b plays an oncogenic role by
repressing PTEN expression in esophageal squamous cell carcinoma
cells. BMC Cancer. 15:292015. View Article : Google Scholar : PubMed/NCBI
|