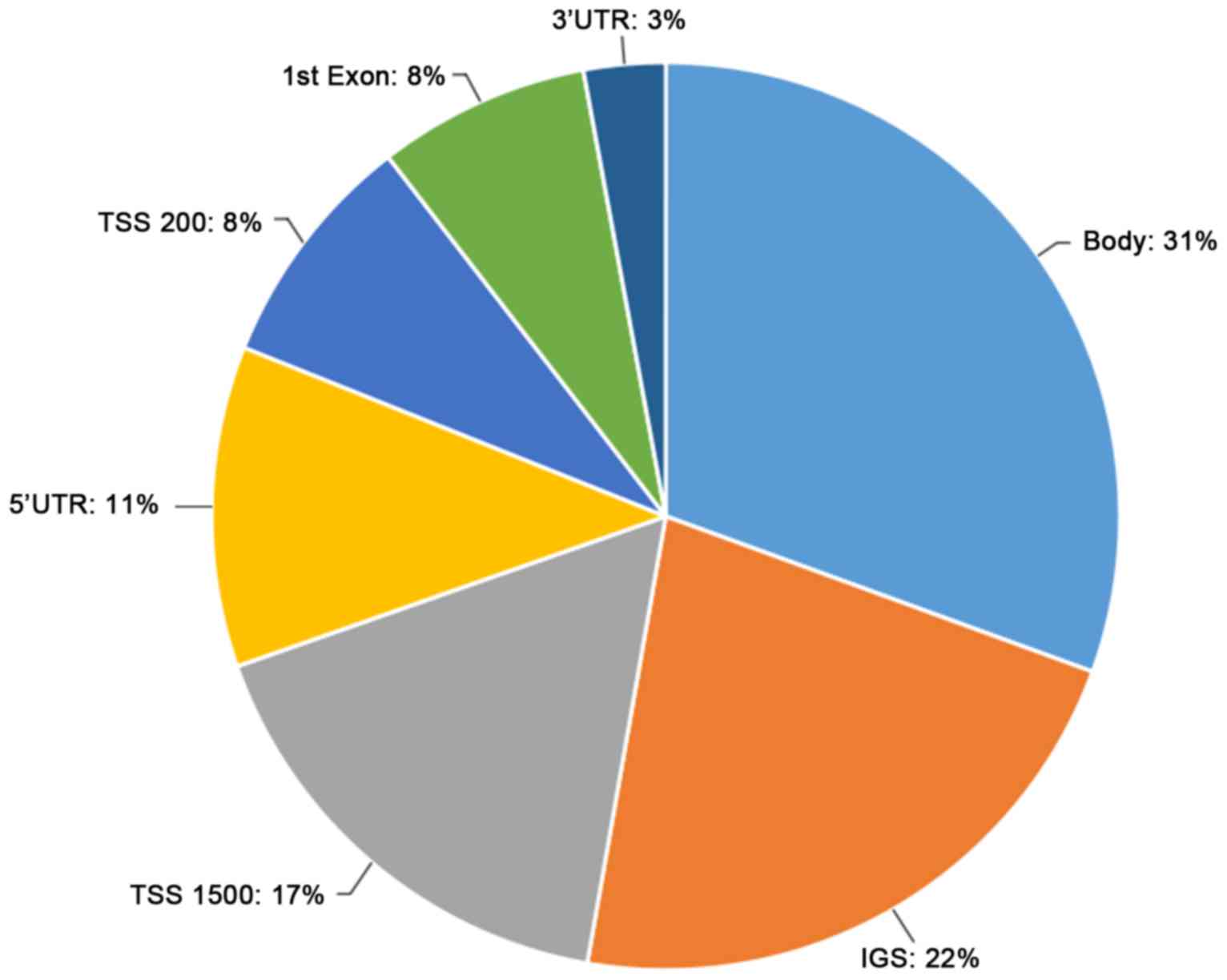
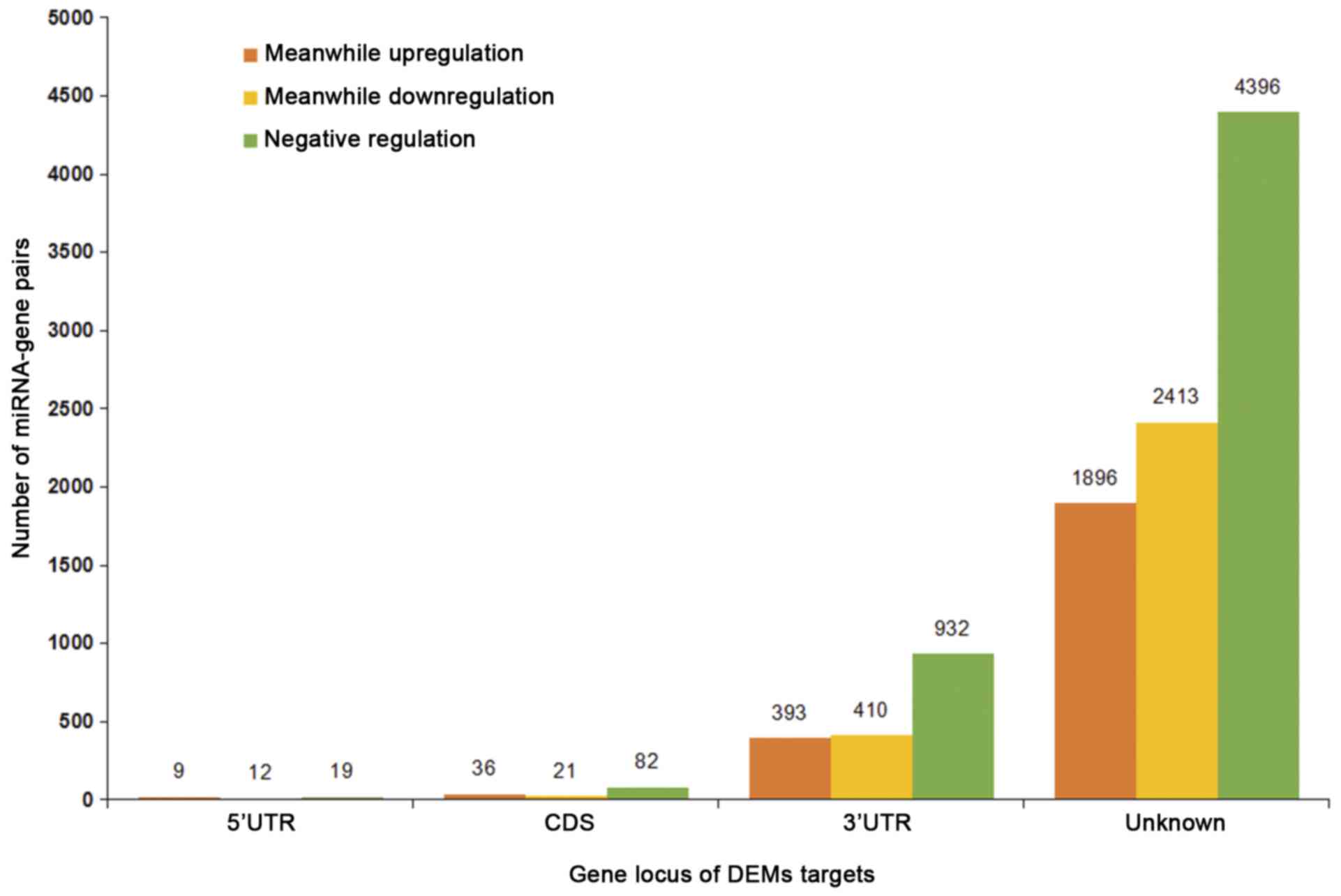
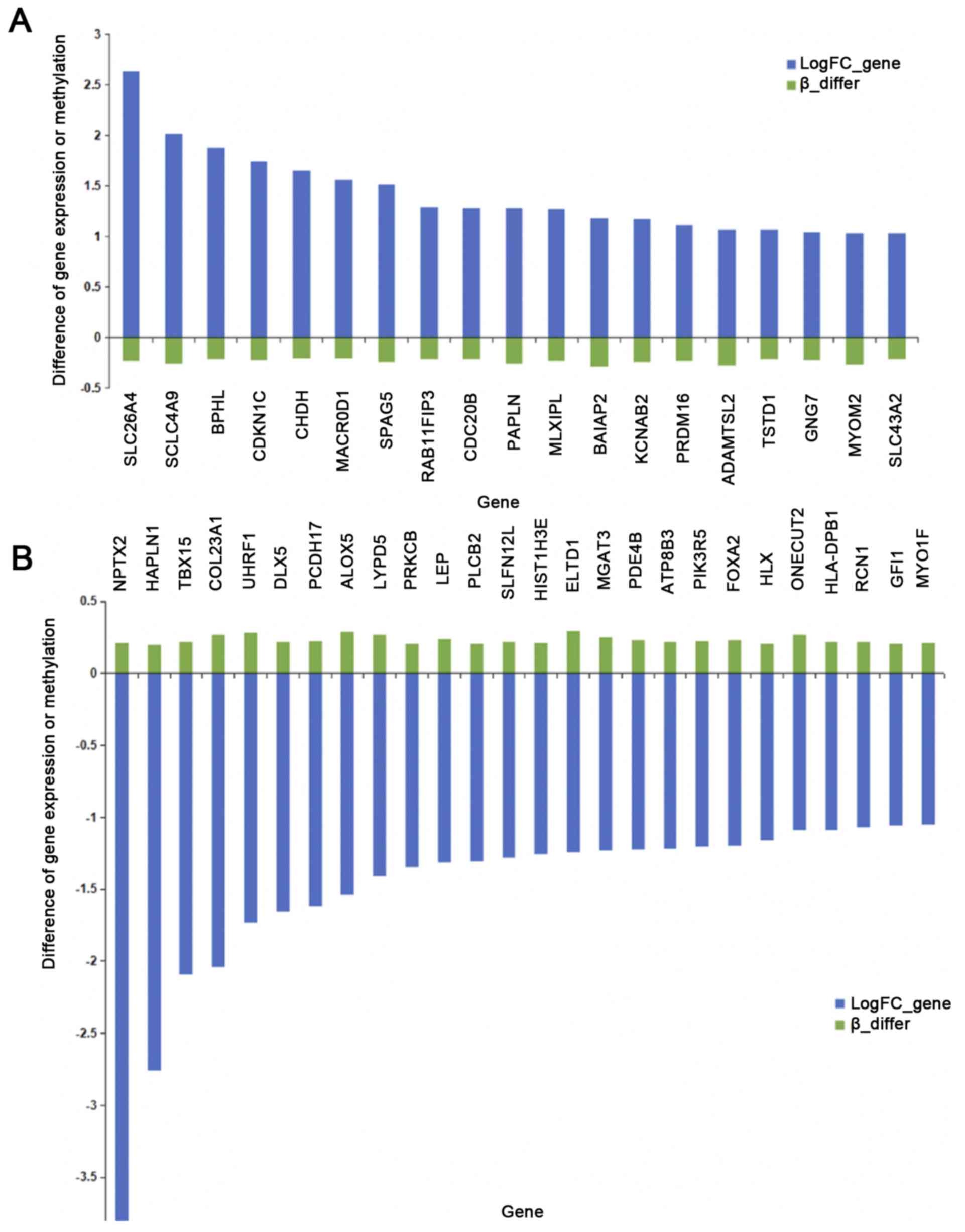
|
1
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Protzel C, Maruschke M and Hakenberg OW:
Epidemiology, aetiology, and pathogenesis of renal cell carcinoma.
Eur Urol Supp. 11:52–59. 2012. View Article : Google Scholar
|
|
3
|
Baek M, Jung JY, Kim JJ, Park KH and Ryu
DS: Characteristics and clinical outcomes of renal cell carcinoma
in children: a single center experience. Int J Urol. 17:737–740.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Häggström C, Rapp K, Stocks T, Manjer J,
Bjørge T, Ulmer H, Engeland A, Almqvist M, Concin H, Selmer R, et
al: Correction: Metabolic factors associated with risk of renal
cell carcinoma. PloS One. 8:e574752013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Trpkov K, Hes O, Agaimy A, Bonert M,
Martinek P, Magi-Galluzzi C, Kristiansen G, Lüders C, Nesi G,
Compérat E, et al: Fumarate hydratase-deficient renal cell
carcinoma is strongly correlated with fumarate hydratase mutation
and hereditary leiomyomatosis and renal cell carcinoma syndrome. Am
J Surg Pathol. 40:8652016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Matsuura K, Nakada C, Mashio M, Narimatsu
T, Yoshimoto T, Tanigawa M, Tsukamoto Y, Hijiya N, Takeuchi I,
Nomura T, et al: Downregulation of SAV1 plays a role in
pathogenesis of high-grade clear cell renal cell carcinoma. BMC
Cancer. 11:5232011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
White NM, Bao TT, Grigull J, Youssef YM,
Girgis A, Diamandis M, Fatoohi E, Metias M, Honey RJ, Stewart R, et
al: miRNA profiling for clear cell renal cell carcinoma: Biomarker
discovery and identification of potential controls and consequences
of miRNA dysregulation. J Urol. 186:1077–1083. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fedorko M, Pacik D, Wasserbauer R, Juracek
J, Varga G, Ghazal M and Nussir MI: microRNAs in the pathogenesis
of renal cell carcinoma and their diagnostic and prognostic utility
as cancer biomarkers. Int J Biol Markers. 31:e26–e37. 2015.
View Article : Google Scholar
|
|
9
|
Tsai HC and Baylin SB: Cancer epigenetics:
Linking basic biology to clinical medicine. Cell Res. 21:502–517.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ricketts CJ, Morris MR, Gentle D, Brown M,
Wake N, Woodward ER, Clarke N, Latif F and Maher ER: Genome-wide
CpG island methylation analysis implicates novel genes in the
pathogenesis of renal cell carcinoma. Epigenetics. 7:278–290. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang X, Chen X, Han W, Ruan A, Chen L,
Wang R, Xu Z, Xiao P3, Lu X, Zhao Y, et al: miR-200c targets CDK2
and suppresses tumorigenesis in renal cell carcinoma. Mol Cancer
Res. 13:1567–1577. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wei JH, Haddad A, Wu KJ, Zhao HW, Kapur P,
Zhang ZL, Zhao LY, Chen ZH, Zhou YY, Zhou JC, et al: A
CpG-methylation-based assay to predict survival in clear cell renal
cell carcinoma. Nat Commun. 6:86992015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wiberg AO, Liu L, Tong Z, Myslivets E,
Ataie V, Kuo BP, Alic N and Radic S: Photonic preprocessor for
analog-to-digital-converter using a cavity-less pulse source. Opt
Express. 20:B419–B427. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nautiyal S, Carlton VE, Lu Y, Ireland JS,
Flaucher D, Moorhead M, Gray JW, Spellman P, Mindrinos M, Berg P
and Faham M: High-throughput method for analyzing methylation of
CpGs in targeted genomic regions. Proc Natl Acad Sci USA.
107:12587–12592. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao YF, Shu Y, Yang L, He YC, Li LP, Huang
G, Li HP and Jiang Y: A graphic method for identification of novel
glioma related genes. Biomed Res Int. 2014:8919452014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Denham J, O'Brien BJ, Harvey JT and
Charchar FJ: Genome-wide sperm DNA methylation changes after 3
months of exercise training in humans. Epigenomics. 7:717–731.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ueno S, Saito S, Wada T, Yamaguchi K,
Satoh M, Arai Y and Miyagi T: Plasma membrane-associated sialidase
is up-regulated in renal cell carcinoma and promotes
interleukin-6-induced apoptosis suppression and cell motility. J
Biol Chem. 281:7756–7764. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tringali C, Lupo B, Silvestri I, Papini N,
Anastasia L, Tettamanti G and Venerando B: The plasma membrane
sialidase NEU3 regulates the malignancy of renal carcinoma cells by
controlling β1 integrin internalization and recycling. J Biol Chem.
287:42835–42845. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Eto M, Harano M, Tatsugami K, Harada M,
Kamiryo Y, Kiyoshima K, Hamaguchi M, Tsuneyoshi M, Yoshikai Y and
Naito S: Cyclophosphamide-using nonmyeloablative allogeneic cell
therapy against renal cancer with a reduced risk of
graft-versus-host disease. Clin Cancer Res. 13:1029–1035. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
van Bergen CA, Verdegaal EME, Honders MW,
Hoogstraten C, Steijn-van Tol AQ, de Quartel L, de Jong J, Meyering
M, Falkenburg JH, Griffioen M and Osanto S: Durable remission of
renal cell carcinoma in conjuncture with graft versus host disease
following allogeneic stem cell transplantation and donor lymphocyte
infusion: Rule or exception? PloS One. 9:e851982014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Massenkeil G, Roigas J, Nagy M, Wille A,
Stroszczynski C, Mapara MY, Loening S, Dörken B and Arnold R:
Nonmyeloablative stem cell transplantation in metastatic renal cell
carcinoma: Delayed graft-versus-tumor effect is associated with
chimerism conversion but transplantation has high toxicity. Bone
Marrow Transplant. 34:309–316. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ahanda Endale ML, Fritz ER, Estellé J, Hu
ZL, Madsen O, Groenen MA, Beraldi D, Kapetanovic R, Hume DA,
Rowland RR, et al: Prediction of altered 3′-UTR miRNA-binding sites
from RNA-Seq data: The swine leukocyte antigen complex (SLA) as a
model region. Plos One. 7:e486072012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ivanova AV, Goparaju CM, Ivanov SV, Nonaka
D, Cruz C, Beck A, Lonardo F, Wali A and Pass HI: Protumorigenic
role of HAPLN1 and its IgV domain in malignant pleural
mesothelioma. Clin Cancer Res. 15:2602–2611. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yau C, Esserman L, Moore DH, Waldman F,
Sninsky J and Benz CC: A multigene predictor of metastatic outcome
in early stage hormone receptor-negative and triple-negative breast
cancer. Breast Cancer Res. 12:R852010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mebarki S, Désert R, Sulpice L, Sicard M,
Desille M, Canal F, Schneider Dubois-Pot H, Bergeat D, Turlin B,
Bellaud P, et al: De novo HAPLN1 expression hallmarks Wnt-induced
stem cell and fibrogenic networks leading to aggressive human
hepatocellular carcinomas. Oncotarget. 7:39026–39043. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zanette DL, Rivadavia F, Molfetta GA,
Barbuzano FG, Proto-Siqueira R, Silva WA Jr, Falcão RP and Zago MA:
miRNA expression profiles in chronic lymphocytic and acute
lymphocytic leukemia. Braz J Med Biol Res. 40:1435–1440. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang Z, Miao R, Li G, Wu Y, Robson SC,
Yang X, Zhao Y, Zhao H and Zhong Y: Identification of recurrence
related microRNAs in hepatocellular carcinoma after surgical
resection. Int J Mol Sci. 14:1105–1118. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang XW, Wu Y, Wang D and Qin ZF: microRNA
network analysis identifies key microRNAs and genes associated with
precancerous lesions of gastric cancer. Genet Mol Res.
13:8695–8703. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jiang B, Liu GW and Xie HH: Expression and
its clinical significance of hsa-miR-218 in tissues of colon
carcinoma. Med Info. 2010.
|
|
33
|
Zhang XL, Shi HJ, Wang JP, Tang HS and Cui
SZ: miR-218 inhibits multidrug resistance (MDR) of gastric cancer
cells by targeting hedgehog/smoothened. Int J Clin Exp Pathol.
8:6397–6406. 2015.PubMed/NCBI
|