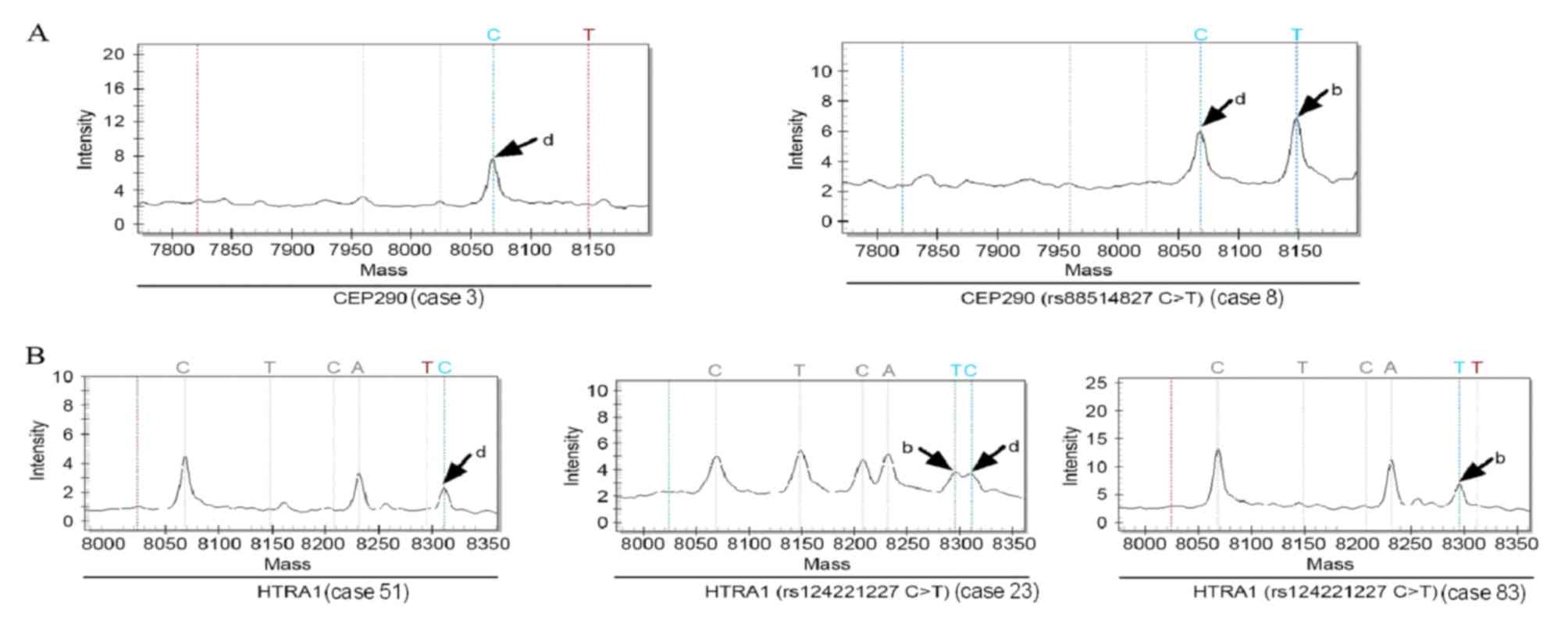
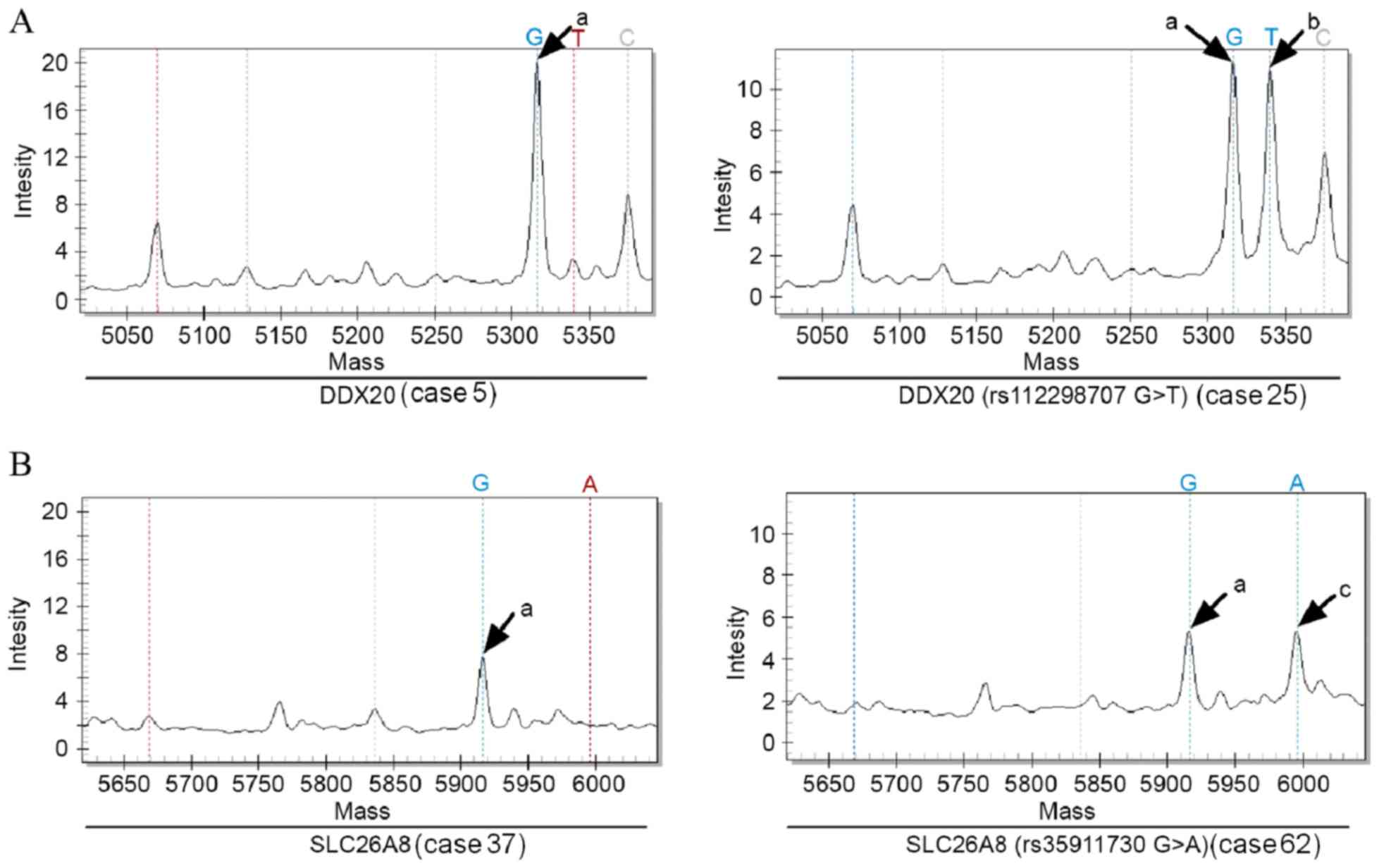
|
1
|
Kastrinos F and Stoffel EM: History,
genetics, and strategies for cancer prevention in Lynch syndrome.
Clin Gastroenterol Hepatol. 12:715–727; quiz e41-43. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Watson P and Riley B: The tumor spectrum
in the Lynch syndrome. Fam Cancer. 4:245–248. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fishel R, Lescoe MK, Rao MR, Copeland NG,
Jenkins NA, Garber J, Kane M and Kolodner R: The human mutator gene
homolog MSH2 and its association with hereditary nonpolyposis colon
cancer. Cell. 75:1027–1038. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bronner CE, Baker SM, Morrison PT, Warren
G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A,
et al: Mutation in the DNA mismatch repair gene homologue hMLH1 is
associated with hereditary non-polyposis colon cancer. Nature.
368:258–261. 1994. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Whitehouse A, Meredith DM and Markham AF:
DNA mismatch repair genes and their association with colorectal
cancer (Review). Int J Mol Med. 1:469–474. 1998.PubMed/NCBI
|
|
6
|
Hampel H, Frankel WL, Martin E, Arnold M,
Khanduja K, Kuebler P, Nakagawa H, Sotamaa K, Prior TW, Westman J,
et al: Screening for the Lynch syndrome (hereditary nonpolyposis
colorectal cancer). N Engl J Med. 352:1851–1860. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lynch HT and de la Chapelle A: Hereditary
colorectal cancer. N Engl J Med. 348:919–932. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bashyam MD, Kotapalli V, Raman R,
Chaudhary AK, Yadav BK, Gowrishankar S, Uppin SG, Kongara R, Sastry
RA, Vamsy M, et al: Evidence for presence of mismatch repair gene
expression positive Lynch syndrome cases in India. Mol Carcinog.
54:1807–1814. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vasen HF, Mecklin JP, Khan PM and Lynch
HT: The International Collaborative Group on Hereditary
Non-Polyposis Colorectal Cancer (ICG-HNPCC). Dis Colon Rectum.
34:424–425. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peltomäki P and Vasen H: Mutations
associated with HNPCC predisposition-Update of ICG-HNPCC/INSiGHT
mutation database. Dis Markers. 20:269–276. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lindor NM: Familial colorectal cancer type
X: The other half of hereditary nonpolyposis colon cancer syndrome.
Surg Oncol Clin N Am. 18:637–645. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nieminen TT, O'Donohue MF, Wu Y, Lohi H,
Scherer SW, Paterson AD, Ellonen P, Abdel-Rahman WM, Valo S,
Mecklin JP, et al: Germline mutation of RPS20, encoding a ribosomal
protein, causes predisposition to hereditary nonpolyposis
colorectal carcinoma without DNA mismatch repair deficiency.
Gastroenterology. 147:595–598.e5. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang H, Dinney CP, Ye Y, Zhu Y, Grossman
HB and Wu X: Evaluation of genetic variants in microRNA-related
genes and risk of bladder cancer. Cancer Res. 68:2530–2537. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shin EM, Hay HS, Lee MH, Goh JN, Tan TZ,
Sen YP, Lim SW, Yousef EM, Ong HT, Thike AA, et al: DEAD-box
helicase DP103 defines metastatic potential of human breast
cancers. J Clin Invest. 124:3807–3824. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sagona AP, Nezis IP, Bache KG, Haglund K,
Bakken AC, Skotheim RI and Stenmark H: A tumor-associated mutation
of FYVE-CENT prevents its interaction with Beclin 1 and interferes
with cytokinesis. PLoS One. 6:e170862011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wen JF BB, Zhang XB and Jiang-Ping XU:
Expression and significance of ZFYVE26 in hepatocellular carcinoma.
J Prac Med. 28:1939–1942. 2012.
|
|
17
|
Cao G, Dong W, Meng X, Liu H, Liao H and
Liu S: MiR-511 inhibits growth and metastasis of human
hepatocellular carcinoma cells by targeting PIK3R3. Tumour Biol.
36:4453–4459. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang G, Yang X, Li C, Cao X, Luo X and Hu
J: PIK3R3 induces epithelial-to-mesenchymal transition and promotes
metastasis in colorectal cancer. Mol Cancer Ther. 13:1837–1847.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wong TS, Gao W and Chan JY: Transcription
regulation of E-cadherin by zinc finger E-box binding homeobox
proteins in solid tumors. Biomed Res Int. 2014:9215642014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shahbazi J, Lock R and Liu T: Tumor
protein 53-induced nuclear protein 1 enhances p53 function and
represses tumorigenesis. Front Genet. 4:802013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou X, Ma L, Li J, Gu J, Shi Q and Yu R:
Effects of SEMA3G on migration and invasion of glioma cells. Oncol
Rep. 28:269–275. 2012.PubMed/NCBI
|
|
22
|
Valletta D, Czech B, Spruss T, Ikenberg K,
Wild P, Hartmann A, Weiss TS, Oefner PJ, Müller M, Bosserhoff AK
and Hellerbrand C: Regulation and function of the atypical cadherin
FAT1 in hepatocellular carcinoma. Carcinogenesis. 35:1407–1415.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Morris LG, Kaufman AM, Gong Y, Ramaswami
D, Walsh LA, Turcan Ş, Eng S, Kannan K, Zou Y, Peng L, et al:
Recurrent somatic mutation of FAT1 in multiple human cancers leads
to aberrant Wnt activation. Nat Genet. 45:253–261. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang JW, Gamsby JJ, Highfill SL, Mora LB,
Bloom GC, Yeatman TJ, Pan TC, Ramne AL, Chodosh LA, Cress WD, et
al: Deregulated expression of LRBA facilitates cancer cell growth.
Oncogene. 23:4089–4097. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Herold T, Metzeler KH, Vosberg S, Hartmann
L, Röllig C, Stölzel F, Schneider S, Hubmann M, Zellmeier E,
Ksienzyk B, et al: Isolated trisomy 13 defines a homogeneous AML
subgroup with high frequency of mutations in spliceosome genes and
poor prognosis. Blood. 124:1304–1311. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu DI, Liu L, Ren C, Kong D, Zhang P, Jin
X, Wang T and Zhang G: Epithelial-mesenchymal interconversions and
the regulatory function of the ZEB family during the development
and progression of ovarian cancer. Oncol Lett. 11:1463–1468. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Childs EJ, Mocci E, Campa D, Bracci PM,
Gallinger S, Goggins M, Li D, Neale RE, Olson SH, Scelo G, et al:
Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with
susceptibility to pancreatic cancer. Nat Genet. 47:911–916. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Archer NS, Nassif NT and O'Brien BA:
Genetic variants of SLC11A1 are associated with both autoimmune and
infectious diseases: Systematic review and meta-analysis. Genes
Immun. 16:275–283. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lohi H, Kujala M, Makela S, Lehtonen E,
Kestila M, Saarialho-Kere U, Markovich D and Kere J: Functional
characterization of three novel tissue-specific anion exchangers
SLC26A7, -A8 and -A9. J Biol Chem. 277:14246–14254. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu F, Jin L, Luo TP, Luo GH, Tan Y and
Qin XH: Serine protease HtrA1 expression in human hepatocellular
carcinoma. Hepatobiliary Pancreat Dis Int. 9:508–512.
2010.PubMed/NCBI
|
|
31
|
Lehner A, Magdolen V, Schuster T, Kotzsch
M, Kiechle M, Meindl A, Sweep FC, Span PN and Gross E:
Downregulation of serine protease HTRA1 is associated with poor
survival in breast cancer. PLoS One. 8:e603592013. View Article : Google Scholar : PubMed/NCBI
|