Introduction
Cervical cancer is the second most common type of
cancer in females worldwide, with an estimated 530,000 novel cases
and 275,000 mortalities annually (1).
The occurrence and development of cervical cancer involves a
sequential progression from normal cervical epithelium to
preneoplastic cervical intraepithelial neoplasia and subsequently
to invasive cervical cancer (2).
Studies have indicated that numerous risk factors contribute to the
initiation and development of cervical cancer including early
sexual intercourse, increased number of sexual partners and
infection with high risk types of human papillomavirus (3,4). Surgery
with adjuvant chemotherapy is the current therapeutic intervention
for patients with cervical cancer. Despite marked progress in the
diagnosis, surgical methods and application of comprehensive
therapy, prognosis remains unsatisfactory (5). The 5-year overall survival rate for
patients with locally advanced cervical cancer is between 30 and
50% compared with between 5 and 15% for patients with metastatic
disease (6). Therefore, it is
imperative to fully understand the underlying molecular mechanism
of cervical cancer and provide novel diagnostic and prognostic
markers, and identify novel therapeutic targets for treatment of
this disease.
MicroRNAs (miRs) are endogenously expressed RNAs of
between 20 and 23 nucleotides in length, which comprise a large
family of non-coding and single-strand RNAs (7). These small molecules have been
demonstrated to regulate gene expression at the
post-transcriptional level through binding to the 3′-untranslated
regions (3′-UTRs) of their target genes in a base-pairing manner,
resulting in translational inhibition or mRNA degradation (8,9). The
association between miRs and cancer has become a popular area of
research. Comparison between human cancer tissues and their matched
normal tissues have revealed distinct miR expression profiles
(10). The abnormal expression of
miRs in various types of cancer is associated with a broad range of
physiological and pathological processes including proliferation,
apoptosis, cell cycle, migration, invasion and metastasis (11,12). In
human cancer, miRs may act as tumor suppressors or oncogenes which
primarily depends on the roles of their target genes (13). Highly expressed miRs function as
oncogenes by blocking tumor suppressor gene function, whereas
downregulated miRs function as tumor suppressor genes through
negative regulation of oncogenes (14,15).
Therefore, elucidating the expression pattern, functional roles and
underlying molecular mechanism of miRs may be particularly useful
in improving cancer treatments.
In the present study, the expression level of
miR-197 was determined in cervical cancer, and the functional roles
of miR-197 were investigated in cervical cancer cells and the
direct target genes of miR-197 were identified in cervical cancer.
The results of the present study determined the differential
expression of miR-197 in cervical cancer and demonstrated that
miR-197 suppressed cell proliferation and invasion in cervical
cancer though directly targeting forkhead box M1 (FOXM1).
Materials and methods
Ethical statement and human tissue
samples
The present study was approved by the Ethics
Committee of Xiangyang Central Hospital (Xiangyang, China). Written
informed consent was obtained from all patients prior to collection
of tissue samples. A total of 46 pairs of human cervical cancer and
adjacent normal cervical tissues were obtained from patients with
cervical cancer who underwent cervical surgical resection without
radiotherapy and/or chemotherapy treatment at Xiangyang Central
Hospital. The tissue samples were frozen in liquid nitrogen
immediately following surgery and stored at −80°C until use.
Cell lines, culture conditions and
cell transfection
The normal human cervical epithelial cell line
(Ect1/E6E7), cervical cancer cell lines (HeLa, C33A, CaSki and
SiHa) and HEK293 T cell line were purchased from Chinese Center for
Type Culture Collection (Wuhan, China). All cell lines were
cultured in Dulbecco's modified Eagle's medium (DMEM) containing
10% (FBS) and 1% antibiotic/antimycotic (all from Thermo Fisher
Scientific, Waltham, MA, USA) at 37°C in a humidified atmosphere
containing 5% CO2. Chemically synthesized miR-197
mimics, negative control (NC), FOXM1 small interfering RNA (siRNA)
and NC siRNA were obtained from Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). The miR-197 mimic sequence was
5′-UUCACCACCUUCUCCACCCCAGC-3′ and the NC sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. The FOXM1 siRNA sequence was
5′-GGACCACUUUCCCUACUUUTT-3′ and the NC siRNA sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. Cells were transfected with miR-197
mimics, NC, FOXM1 siRNA or NC siRNA using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from tissues and cells were isolated using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. To determine the levels
of miR-197, RT was performed using the miScript Reverse
Transcription kit (Qiagen China Co., Ltd., Shanghai, China),
followed by qPCR using SYBR Premix Ex Taq II (Takara Biotechnology
Co., Ltd., Dalian, China). The qPCR was performed with cycling
conditions as follows: 5 min at 95°C, followed by 40 cycles of 95°C
for 30 sec and 65°C for 45 sec. To monitor FOXM1 expression, cDNA
was synthesized from total RNA using Moloney murine leukemia virus
reverse transcriptase (Invitrogen; Thermo Fisher Scientific. Inc.).
RT-qPCR was performed using a SYBR-Green Master Mix kit (Roche
Applied Science, Shanghai, China). The thermocycling conditions for
qPCR were as follows: 95°C for 10 min, followed by 40 cycles of
95°C for 15 sec and 60°C for 1 min. The primers were designed as
follows: miR-197, 5′-ATTACTTTGCCCATATTCATTTTGA-3′ (forward) and
5′-ATTCTAGAGGCCGAGGCGGCCGACATGT-3′ (reverse); U6,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and
5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); FOXM1,
5′-GAAGAACTCCATCCGCCACA-3′ (forward) and
5′-GCCTTAAACACCTGGTCCAATGTC-3′ (reverse); and β-actin,
5′-TGGCATTGTTACCAACTGGGTC-3′ (forward) and
5′-TCACGGTTGGCCTTAGGGTTC-3′ (reverse). Relative expression of
miR-197 and FOXM1 mRNA was normalized to U6 and β-actin,
respectively. Relative expression was calculated using the
2−ΔΔCq method (16).
MTT assay
Cells were seeded separately in 96-well plates at a
density of 3,000 cells/well 24 h after transfection. The MTT (5
mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) assay was
performed 0, 24, 48, 72 and 96 h after incubation. In brief, 10 µl
MTT solution was added to each well and the plates were then
incubated for an additional 4 h at 37°C. Subsequently, the culture
medium was removed, 200 µl dimethyl sulfoxide (Sigma-Aldrich; Merck
KGaA) was added to each well and the plates were incubated at 37°C
for 30 min. Finally, the optical density at 490 nm was measured
using a Versamax microplate reader (Molecular Devices, LCC,
Sunnyvale, CA, USA).
In vitro cell invasion assay
A 24-well Boyden chamber with an 8-µm pore-size
polycarbonate membrane (Corning Life Sciences, Cambridge, MA, USA)
was used to evaluate the invasion ability of cervical cancer cells.
The membranes were coated with Matrigel (BD Biosciences, San Jose,
CA, USA) according to the manufacturer's protocol. Cells
(5×104) were collected and reseeded into the upper
chamber in 200 µl FBS-free culture medium (Gibco; Thermo Fisher
Scientific, Inc.) 48 h after transfection. In the lower chamber,
500 µl DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 20%
FBS was added as a chemoattractant. Following incubation for 48 h,
cells remaining on the top of the membranes were carefully removed.
Then cells migrating across the membranes were fixed, stained with
0.1% crystal violet (Beyotime Institute of Biotechnology, Haimen,
China) at room temperature for 20 min and counted under a light
microscope in 5 fields.
Target predication and luciferase
reporter assay
To explore the potential target genes of miR-197,
bioinformatic analysis was performed using miRanda (www.microrna.org/microrna) and TargetScan
(www.targetscan.org).
To explore the direct interaction between miR-197
and FOXM1, a luciferase reporter assay was performed. HEK293 T
cells were seeded in 24-well plates at a density of between 40 and
50% confluence and transfected with miR-197 mimics or NC, along
with pGL3-FOXM1-3′UTR wild-type (Wt) or pGL3-FOXM1-3′UTR mutant
(Mut). pGL3-FOXM1-3′UTR Wt and pGL3-FOXM1-3′UTR Mut luciferase
reporter vectors, synthesized by Shanghai GenePharma Co., Ltd.
(Shanghai, China). The luciferase activity was measured using
Dual-Luciferase Reporter assays (Promega Corporation, Madison, WI,
USA) 48 h post-transfection and the transfection efficiency was
normalized to the paired Renilla luciferase activity.
Results were obtained from three independent experiments.
Western blotting
Proteins were isolated from transfected cells using
radioimmunoprecipitation lysis buffer (Beyotime Institute of
Biotechnology) in the presence of protease inhibitor cocktail (0.1
mg/ml phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate and
1 mg/ml aprotinin; Pierce; Thermo Fisher Scientific, Inc.).
Following centrifugation at 10,000 × g for 15 min, supernatants
were transferred to new tubes for quantification. The concentration
of total protein was determined with bicinchoninic acid protein
assay (Aidlab Biotechnologies Co., Ltd., Beijing, China). Equal
amounts of protein (20 µg) were separated using SDS-PAGE (10% gel),
transferred onto polyvinylidene difluoride membranes (EMD
Millipore, Billerica, MA, USA) and blocked with 5% non-fat milk in
Tris-buffered saline-Tween-20 (TBST) buffer for 0.5 h at room
temperature. The membranes were then probed overnight at 4°C with
mouse anti-human monoclonal FOXM1 (1:1,000 dilution, sc-166709;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and GADPH (1:1,000
dilution, sc-59540; Santa Cruz Biotechnology, Inc.) antibodies.
Following washing three times with TBST, the membranes were
incubated with a goat anti-mouse horseradish peroxidase-conjugated
secondary antibody (Santa Cruz Biotechnology, Inc.) for 2 h at room
temperature. Finally, an enhanced chemiluminescent reagent (EMD
Millipore) was added to develop the signal bands. The intensity of
signal bands was analyzed with GeneTools (version 3.03; SynGene,
Frederick, MD, USA).
Statistical analysis
All values are presented as the mean ± standard
deviation. Differences between groups were assessed using SPSS
software (version 13.0; SPSS, Inc., Chicago, IL, USA). Data were
analyzed with Student's t-test or one-way analysis of variance.
Students-Newman-Keuls was performed to compare between two groups
in multiple groups study. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-197 downregulation in cervical
cancer tissues and cell lines
The expression level of miR-197 was measured in 46
pairs of cervical cancer and adjacent normal cervical tissues using
RT-qPCR. It was identified that miR-197 was significantly
downregulated in cervical cancer tissues compared with adjacent
normal cervical tissues (P<0.05; Fig.
1A). miR-197 expression in human cervical cancer cell lines
(HeLa, C33A, CaSki and SiHa) and a normal human cervical epithelial
cell line (Ect1/E6E7) was also detected. As presented in Fig. 1B, the expression levels of miR-197
were significantly decreased in the four cervical cancer cell lines
compared with Ect1/E6E7 cells (P<0.05).
Effects of miR-197 on viability and
invasion of cervical cancer cells
To investigate the roles of miR-197 in cervical
cancer, HeLa and SiHa cells expressing a relatively low level of
miR-197 were transfected with miR-197 mimics or NC. Following
transfection, the expression level of miR-197 was significantly
increased in HeLa and SiHa cells transfected with miR-197 mimics
(P<0.05; Fig. 2A).
MTT assays were performed to investigate the effect
of miR-197 on the viability of cervical cancer cells.
Overexpression of miR-197 was identified to significantly suppress
the viability of HeLa and SiHa cells (P<0.05; Fig. 2B). To investigate the role of miR-197
on the invasion of the cervical cancer cells, in vitro cell
invasion assays were performed. As shown in Fig. 2C, restoration of miR-197 expression
inhibited the invasive ability of HeLa and SiHa cells compared with
cells transfected with NC (P<0.05). The results of the present
study demonstrated that miR-197 acted as a tumor suppressor in
cervical cancer.
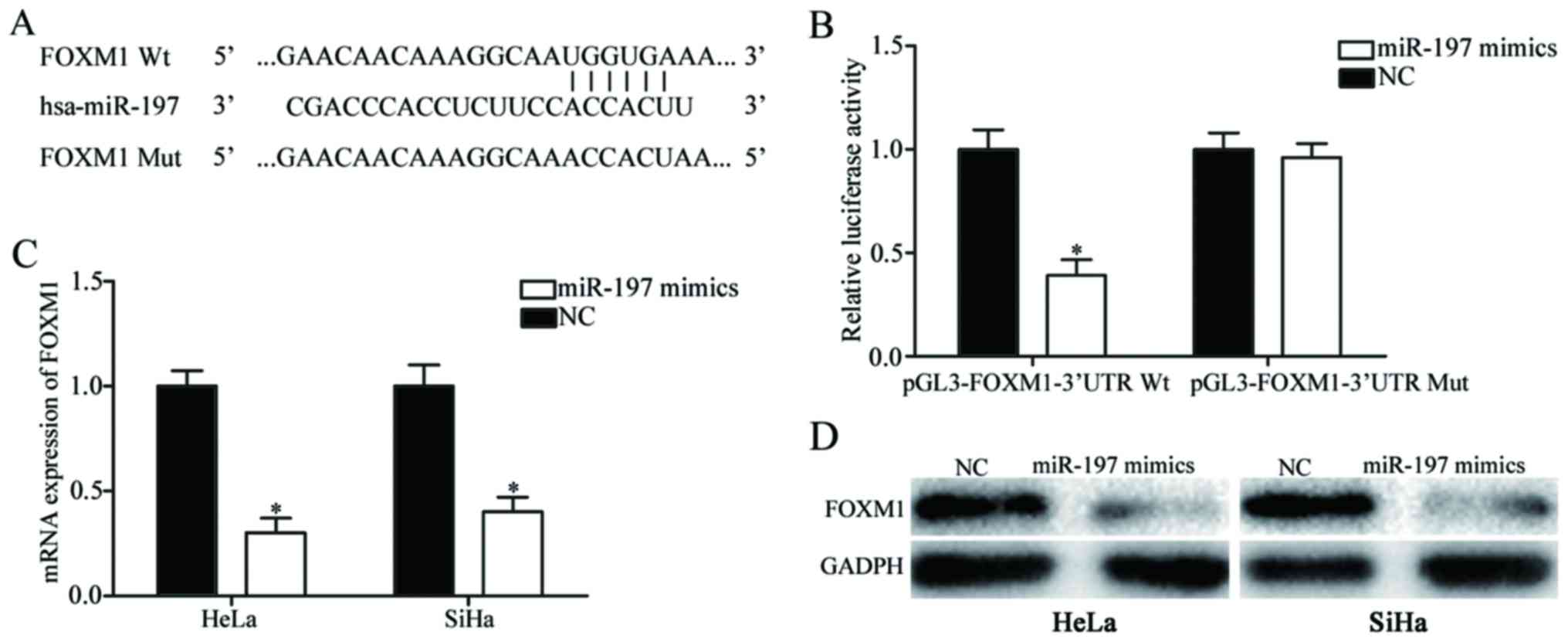
miR-197 directly targets the 3′-UTR of
FOXM1 mRNA to inhibit its expression
To investigate the molecular mechanism underlying
the suppressive roles of miR-197 in cervical cancer, bioinformatic
analysis was performed to explore potential target genes. Among
these putative targets of miR-197, FOXM1 was selected for further
investigation (Fig. 3A). Luciferase
reporter assays revealed that upregulation of miR-197 significantly
decreased the firefly luciferase activity of pGL3-FOXM1-3′UTR Wt
(P<0.05; Fig. 3B), whereas no such
inhibitory effect was identified when miR-197 was transfected with
pGL3-FOXM1-3′UTR Mut (P>0.05).
The effects of miR-197 on FOXM1 expression were also
evaluated. As presented in Fig. 3C and
D, miR-197 overexpression suppressed FOXM1 expression in HeLa
and SiHa cells at the mRNA and protein level (P<0.05). These
results indicated that FOXM1 was a direct target gene of
miR-197.
Effects of FOXM1 silencing on
viability and invasion of cervical cancer cells
To investigate the functional roles of FOXM1 in
cervical cancer, loss-of-function studies using FOXM1 siRNA were
performed. RT-qPCR was performed to evaluate its transfection
efficiency. The results revealed that FOXM1 siRNA significantly
suppressed FOXM1 expression in HeLa and SiHa cells when compared
with cells transfected with NC siRNA (P<0.05; Fig. 4A).
The MTT assays revealed that cell viability was
inhibited in the HeLa and SiHa cells transfected with FOXM1 siRNA
compared with cells transfected with NC siRNA (P<0.05; Fig. 4B). Furthermore, the number of invading
cells was also significantly decreased in FOXM1 siRNA transfectants
compared with NC siRNA transfectants in HeLa and SiHa cells. These
results suggested that the roles of FOXM1 underexpression were
similar with the functions induced by miR-197 overexpression in
cervical cancer cells, suggesting that FOXM1 acted as a downstream
effector in the miR-197-mediated viability and invasion of cervical
cancer cells.
Discussion
It has been demonstrated that miRs serve a critical
role in the carcinogenesis and progression of human cancers
(17,18). Therefore, understanding the roles of
miRs may be important for providing new insights into the involved
molecule mechanism in cancer initiation and development and
defining novel markers for cancer prognosis, diagnosis and
treatment (19). Previously, miR-197
has been reported to be downregulated in glioblastoma (20), uterine leiomyoma (21), multiple myeloma (22) and esophageal cancer (23), and upregulated in non-small cell lung
cancer (24), taxol-resistant ovarian
cancer (25) and hepatocellular
carcinoma (26).
The results of the present study revealed that
miR-197 was downregulated in cervical cancer tissues and cell lines
compared with adjacent normal cervical tissues and normal human
cervix epithelial cell line, respectively. These results
demonstrated that miR-197 may act as a tissue-specific miR.
It has also been reported that miR-197 is associated
with the clinicopathological features in patients with cancer. For
example, in non-small cell lung cancer, miR-197 was associated with
increased tumor size and squamous cell carcinoma histological type.
Furthermore, miR-197 expression was identified as a novel
independent predictor of unfavorable prognosis for patients with
non-small cell lung cancer (24). In
esophageal cancer, results of Kaplan-Meier estimator analysis
suggested the expression levels of miR-197 was markedly associated
with survival time. Furthermore, Cox's multi-factor analysis model
revealed that miR-197 expression was associated with prognosis,
tumor length and expression, and survival time (23). These results indicated that miR-197
may be a biomarker of, and be involved in, the progression of human
cancer.
miR-197 has been reported to be involved in
physiological and pathological processes in numerous types of
cancer. For example, restoration of miR-197 decreased cell
proliferation through negative regulation of Grb-associated-binding
protein 2 (20). In uterine
leiomyoma, upregulation of miR-197 inhibited cellular proliferation
and promoted cell cycle arrest in G0/G1 phase in vitro
(21). Wu et al (27) also demonstrated that miR-197 may
inhibit uterine leiomyoma cell proliferation and migration, and
induce apoptosis in vitro. miR-197 overexpression may
enhance taxol resistance in ovarian cancer cells while also
increasing cell proliferation and invasion (25). Yang et al (22) revealed that miR-197 overexpression
suppressed cell viability, colony formation and migration, and
induced apoptosis in multiple myeloma cells. In hepatocellular
carcinoma, miR-197 underexpression repressed cell migration and
invasion in vitro and in vivo (26). The results of the present study
demonstrated that miR-197 expression decreased cell proliferation
and invasion of cervical cancer cells. These results suggested that
miR-197 acted as a tumor suppressor in cervical cancer and that low
expression level of miR-197 in cervical cancer may contribute to
abnormal proliferation and invasion of cervical cancer cells, and
promote tumor growth and metastasis.
The various effects of miR-197 in distinct tissues
may be a result of the specific targets repressed in each tissue.
In the present study, FOXM1 was identified as a novel target gene
of miR-197. miR-197 target genes in cervical cancer were analyzed
using the TargetScan and miRanda databases. Bioinformatic analysis
results indicated that FOXM1 may be a direct miR-197 target gene.
FOXM1 was selected for further study for the following reasons: i)
FOXM1 is a member of the forkhead superfamily of transcription
factors and has been identified to be upregulated in a number of
types of human cancer including lung, breast, liver, pancreatic and
cervical cancer, as well as in glioblastoma (28–30); and
ii) functional studies have revealed FOXM1 to be involved in
numerous biological processes including cell proliferation, cell
cycle progression, cell differentiation, DNA damage repair, tissue
homeostasis, angiogenesis and apoptosis (31).
Luciferase reporter assays were performed in order
to investigate the hypothesis. The present study revealed that
miR-197 decreased the firefly luciferase activity of a Wt FOXM1
3′-UTR luciferase reporter vector, but did not affect the
luciferase activity of a mut FOXM1 3′-UTR luciferase reporter
vector. Furthermore, restoration of miR-197 expression suppressed
FOXM1 expression at the mRNA and protein levels in cervical cancer
cells. Finally, the roles of FOXM1 underexpression were similar to
the functions induced by miR-197 overexpression in cervical cancer
cells, suggesting that FOXM1 acted as a downstream effector in
miR-197-mediated proliferation and invasion of cervical cancer
cells. The results of the present study demonstrated that miR-197
directly decreased FOXM1 expression by binding to the 3′-UTR of the
FOXM1 gene.
In conclusion, the results of the present study
indicated that miR-197 was downregulated in cervical cancer. In
addition, miR-197 overexpression may suppress cell viability and
invasion of cervical cancer. Furthermore, FOXM1 was identified as a
novel direct target of miR-197. These results suggest the
therapeutic potential of miR-197 in the treatment of cervical
cancer.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paradkar PH, Joshi JV, Mertia PN, Agashe
SV and Vaidya RA: Role of cytokines in genesis, progression and
prognosis of cervical cancer. Asian Pac J Cancer Prev.
15:3851–3864. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bosch FX and de Sanjosé S: Chapter 1:
Human papillomavirus and cervical cancer-burden and assessment of
causality. J Natl Cancer Inst Monogr. 3–13. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yu Y, Zhang Y and Zhang S: MicroRNA-92
regulates cervical tumorigenesis and its expression is upregulated
by human papillomavirus-16 E6 in cervical cancer cells. Oncol Lett.
6:468–474. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Morice P and Castaigne D: Advances in the
surgical management of invasive cervical cancer. Curr Opin Obstet
Gynecol. 17:5–12. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mayr NA, Huang Z, Wang JZ, Lo SS, Fan JM,
Grecula JC, Sammet S, Sammet CL, Jia G, Zhang J, et al:
Characterizing tumor heterogeneity with functional imaging and
quantifying high-risk tumor volume for early prediction of
treatment outcome: Cervical cancer as a model. Int J Radiat Oncol
Biol Phys. 83:972–979. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Carthew RW: Gene regulation by microRNAs.
Curr Opin Genet Dev. 16:203–208. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pereira PM, Marques JP, Soares AR, Carreto
L and Santos MA: MicroRNA expression variability in human cervical
tissues. PLoS One. 5:e117802010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun YC, Wang J, Guo CC, Sai K, Wang J,
Chen FR, Yang QY, Chen YS, Wang J, To TS, et al: MiR-181b
sensitizes glioma cells to teniposide by targeting MDM2. BMC
Cancer. 14:6112014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ventura A and Jacks T: MicroRNAs and
cancer: Short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang T, Thakur A, Chen T, Yang L, Lei G,
Liang Y, Zhang S, Ren H and Chen M: MicroRNA-15a induces cell
apoptosis and inhibits metastasis by targeting BCL2L2 in non-small
cell lung cancer. Tumour Biol. 36:4357–4365. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu J, Valencia-Sanchez MA, Hannon GJ and
Parker R: MicroRNA-dependent localization of targeted mRNAs to
mammalian P-bodies. Nat Cell Biol. 7:719–723. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: microRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tian LQ, Liu EQ, Zhu XD, Wang XG, Li J and
Xu GM: MicroRNA-197 inhibits cell proliferation by targeting GAB2
in glioblastoma. Mol Med Rep. 13:4279–4288. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ling J, Jiang L, Zhang C, Dai J, Wu Q and
Tan J: Upregulation of miR-197 inhibits cell proliferation by
directly targeting IGFBP5 in human uterine leiomyoma cells. In
Vitro Cell Dev Biol Anim. 51:835–842. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang Y, Li F, Saha MN, Abdi J, Qiu L and
Chang H: miR-137 and miR-197 induce apoptosis and suppress
tumorigenicity by targeting MCL-1 in multiple myeloma. Clin Cancer
Res. 21:2399–2411. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang TY, Liu SG, Zhao BS, Qi B, Qin XG and
Yao WJ: Implications of microRNA-197 downregulated expression in
esophageal cancer with poor prognosis. Genet Mol Res. 13:5574–5581.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mavridis K, Gueugnon F, Petit-Courty A,
Courty Y, Barascu A, Guyetant S and Scorilas A: The oncomiR miR-197
is a novel prognostic indicator for non-small cell lung cancer
patients. Br J Cancer. 112:1527–1535. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zou D, Wang D, Li R, Tang Y, Yuan L, Long
X and Zhou Q: MiR-197 induces Taxol resistance in human ovarian
cancer cells by regulating NLK. Tumour Biol. 36:6725–6732. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dai W, Wang C, Wang F, Wang Y, Shen M,
Chen K, Cheng P, Zhang Y, Yang J, Zhu R, et al: Anti-miR-197
inhibits migration in HCC cells by targeting KAI 1/CD82. Biochem
Biophys Res Commun. 446:541–548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu X, Ling J, Fu Z, Ji C, Wu J and Xu Q:
Effects of miRNA-197 overexpression on proliferation, apoptosis and
migration in levonorgestrel treated uterine leiomyoma cells. Biomed
Pharmacother. 71:1–6. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kaestner KH, Knochel W and Martinez DE:
Unified nomenclature for the winged helix/forkhead transcription
factors. Genes Dev. 14:142–146. 2000.PubMed/NCBI
|
|
29
|
Koo CY, Muir KW and Lam EW: FOXM1: From
cancer initiation to progression and treatment. Biochim Biophys
Acta. 1819:28–37. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He SY, Shen HW, Xu L, Zhao XH, Yuan L, Niu
G, You ZS and Yao SZ: FOXM1 promotes tumor cell invasion and
correlates with poor prognosis in early-stage cervical cancer.
Gynecol Oncol. 127:601–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hou Y, Li W, Sheng Y, Li L, Huang Y, Zhang
Z, Zhu T, Peace D, Quigley JG, Wu W, et al: The transcription
factor Foxm1 is essential for the quiescence and maintenance of
hematopoietic stem cells. Nat Immunol. 16:810–818. 2015. View Article : Google Scholar : PubMed/NCBI
|