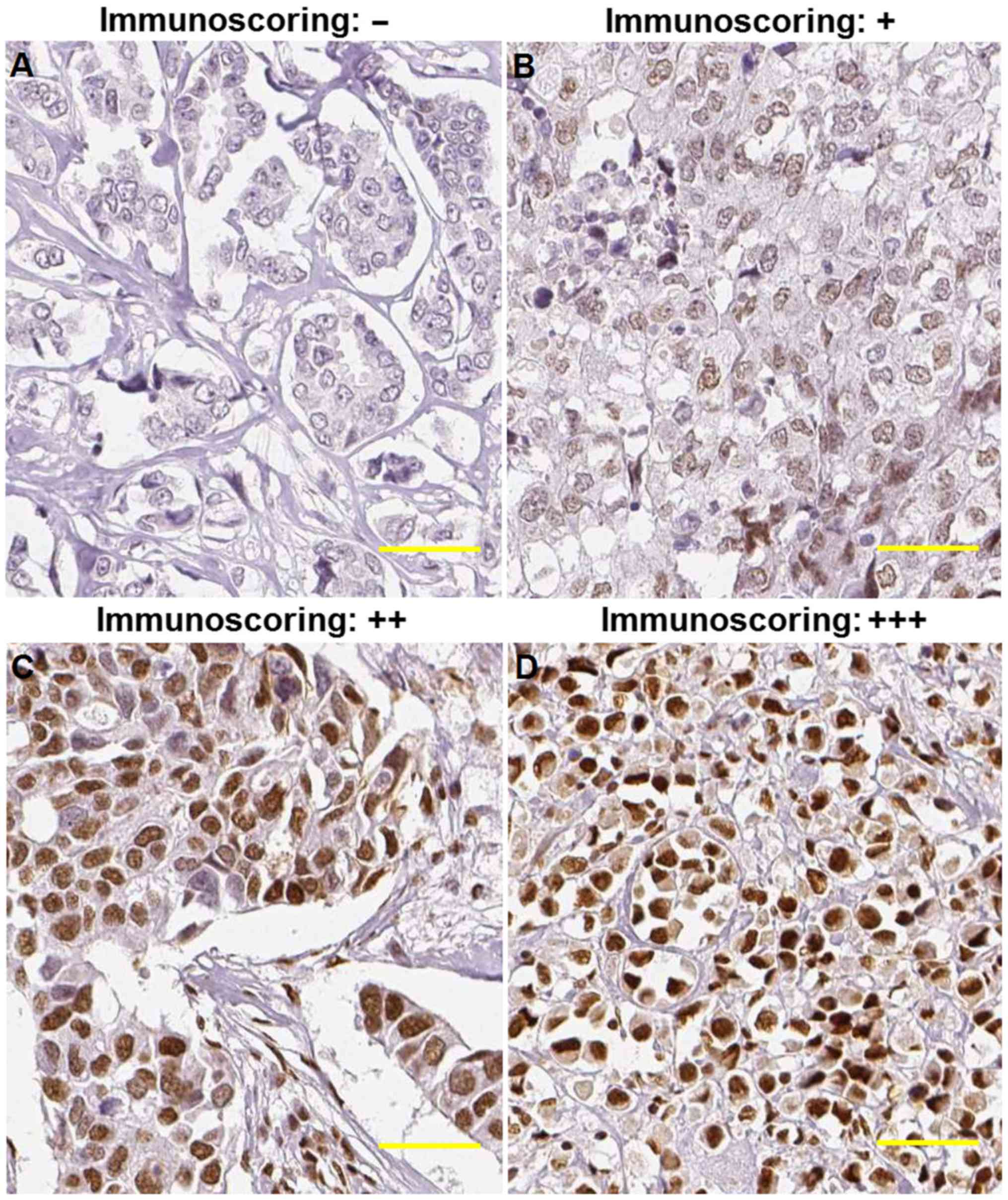
|
1
|
Bozorgi A, Khazaei M and Khazaei MR: New
findings on breast cancer stem cells: A review. J Breast Cancer.
18:303–312. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim SJ, Kim YS, Jang ED, Seo KJ and Kim
JS: Prognostic impact and clinicopathological correlation of CD133
and ALDH1 expression in invasive breast cancer. J Breast Cancer.
18:347–355. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Weigelt B, Peterse JL and van't Veer LJ:
Breast cancer metastasis: Markers and models. Nat Rev Cancer.
5:591–602. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wade MA, Jones D, Wilson L, Stockley J,
Coffey K, Robson CN and Gaughan L: The histone demethylase enzyme
KDM3A is a key estrogen receptor regulator in breast cancer.
Nucleic Acids Res. 43:196–207. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ramadoss S, Guo G and Wang CY: Lysine
demethylase KDM3A regulates breast cancer cell invasion and
apoptosis by targeting histone and the non-histone protein p53.
Oncogene. 36:47–59. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yamane K, Toumazou C, Tsukada Y,
Erdjument-Bromage H, Tempst P, Wong J and Zhang Y: JHDM2A, a
JmjC-containing H3K9 demethylase, facilitates transcription
activation by androgen receptor. Cell. 125:483–495. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wellmann S, Bettkober M, Zelmer A, Seeger
K, Faigle M, Eltzschig HK and Bührer C: Hypoxia upregulates the
histone demethylase JMJD1A via HIF-1. Biochem Biophys Res Commun.
372:892–897. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Parrish JK, Sechler M, Winn RA and
Jedlicka P: The histone demethylase KDM3A is a
microRNA-22-regulated tumor promoter in Ewing Sarcoma. Oncogene.
34:257–262. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cho HS, Toyokawa G, Daigo Y, Hayami S,
Masuda K, Ikawa N, Yamane Y, Maejima K, Tsunoda T, Field HI, et al:
The JmjC domain-containing histone demethylase KDM3A is a positive
regulator of the G1/S transition in cancer cells via
transcriptional regulation of the HOXA1 gene. Int J Cancer.
131:E179–E189. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tee AE, Ling D, Nelson C, Atmadibrata B,
Dinger ME, Xu N, Mizukami T, Liu PY, Liu B, Cheung B, et al: The
histone demethylase JMJD1A induces cell migration and invasion by
up-regulating the expression of the long noncoding RNA MALAT1.
Oncotarget. 5:1793–1804. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yamada D, Kobayashi S, Yamamoto H,
Tomimaru Y, Noda T, Uemura M, Wada H, Marubashi S, Eguchi H,
Tanemura M, et al: Role of the hypoxia-related gene, JMJD1A, in
hepatocellular carcinoma: Clinical impact on recurrence after
hepatic resection. Ann Surg Oncol. 3 Suppl 19:S355–S364. 2012.
View Article : Google Scholar
|
|
14
|
Guo X, Shi M, Sun L, Wang Y, Gui Y, Cai Z
and Duan X: The expression of histone demethylase JMJD1A in renal
cell carcinoma. Neoplasma. 58:153–157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hewitt SM, Baskin DG, Frevert CW, Stahl WL
and Rosa-Molinar E: Controls for immunohistochemistry: The
Histochemical Society's standards of practice for validation of
immunohistochemical assays. J Histochem Cytochem. 62:693–697. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Burry RW: Controls for
immunocytochemistry: An update. J Histochem Cytochem. 59:6–12.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Okada Y, Scott G, Ray MK, Mishina Y and
Zhang Y: Histone demethylase JHDM2A is critical for Tnp1 and Prm1
transcription and spermatogenesis. Nature. 450:119–123. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Beyer S, Kristensen MM, Jensen KS,
Johansen JV and Staller P: The histone demethylases JMJD1A and
JMJD2B are transcriptional targets of hypoxia-inducible factor HIF.
J Biol Chem. 283:36542–36552. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pollard PJ, Loenarz C, Mole DR, McDonough
MA, Gleadle JM, Schofield CJ and Ratcliffe PJ: Regulation of
Jumonji-domain-containing histone demethylases by hypoxia-inducible
factor (HIF)-1alpha. Biochem J. 416:387–394. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou X, Sun H, Ellen TP, Chen H and Costa
M: Arsenite alters global histone H3 methylation. Carcinogenesis.
29:1831–1836. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Krieg AJ, Rankin EB, Chan D, Razorenova O,
Fernandez S and Giaccia AJ: Regulation of the histone demethylase
JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene
expression and tumor growth. Mol Cell Biol. 30:344–353. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Osawa T, Tsuchida R, Muramatsu M,
Shimamura T, Wang F, Suehiro J, Kanki Y, Wada Y, Yuasa Y, Aburatani
H, et al: Inhibition of histone demethylase JMJD1A improves
anti-angiogenic therapy and reduces tumor-associated macrophages.
Cancer Res. 73:3019–3028. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang H, Liu Z, Yuan C, Zhao Y, Wang L, Hu
J, Xie D, Wang L and Chen D: Elevated JMJD1A is a novel predictor
for prognosis and a potential therapeutic target for gastric
cancer. Int J Clin Exp Pathol. 8:11092–11099. 2015.PubMed/NCBI
|
|
24
|
Suikki HE, Kujala PM, Tammela TL, van
Weerden WM, Vessella RL and Visakorpi T: Genetic alterations and
changes in expression of histone demethylases in prostate cancer.
Prostate. 70:889–898. 2010.PubMed/NCBI
|
|
25
|
Zhao QY, Lei PJ, Zhang X, Zheng JY, Wang
HY, Zhao J, Li YM, Ye M, Li L, Wei G and Wu M: Global histone
modification profiling reveals the epigenomic dynamics during
malignant transformation in a four-stage breast cancer model. Clin
Epigenetics. 8:342016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang L, Chang J, Varghese D, Dellinger M,
Kumar S, Best AM, Ruiz J, Bruick R, Peña-Llopis S, Xu J, et al: A
small molecule modulates Jumonji histone demethylase activity and
selectively inhibits cancer growth. Nat Commun. 4:20352013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Baker M: Reproducibility crisis: Blame it
on the antibodies. Nature. 521:274–276. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sar A, Ponjevic D, Nguyen M, Box AH and
Demetrick DJ: Identification and characterization of demethylase
JMJD1A as a gene upregulated in the human cellular response to
hypoxia. Cell Tissue Res. 337:223–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Khouja MH, Baekelandt M, Sarab A, Nesland
JM and Holm R: Limitations of tissue microarrays compared with
whole tissue sections in survival analysis. Oncol Lett. 1:827–831.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Merseburger AS, Kuczyk MA, Serth J,
Bokemeyer C, Young DY, Sun L, Connelly RR, McLeod DG, Mostofi FK,
Srivastava SK, et al: Limitations of tissue microarrays in the
evaluation of focal alterations of bcl-2 and p53 in whole mount
derived prostate tissues. Oncol Rep. 10:223–228. 2003.PubMed/NCBI
|
|
31
|
Mirlacher M, Kasper M, Storz M, Knecht Y,
Dürmüller U, Simon R, Mihatsch MJ and Sauter G: Influence of slide
aging on results of translational research studies using
immunohistochemistry. Mod Pathol. 17:1414–1420. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dressler LG, Geradts J, Burroughs M, Cowan
D, Millikan RC and Newman B: Policy guidelines for the utilization
of formalin-fixed, paraffin-embedded tissue sections: The UNC SPORE
experience. University of North Carolina Specialized Program of
Research Excellence. Breast Cancer Res Treat. 58:31–39. 1999.
View Article : Google Scholar : PubMed/NCBI
|