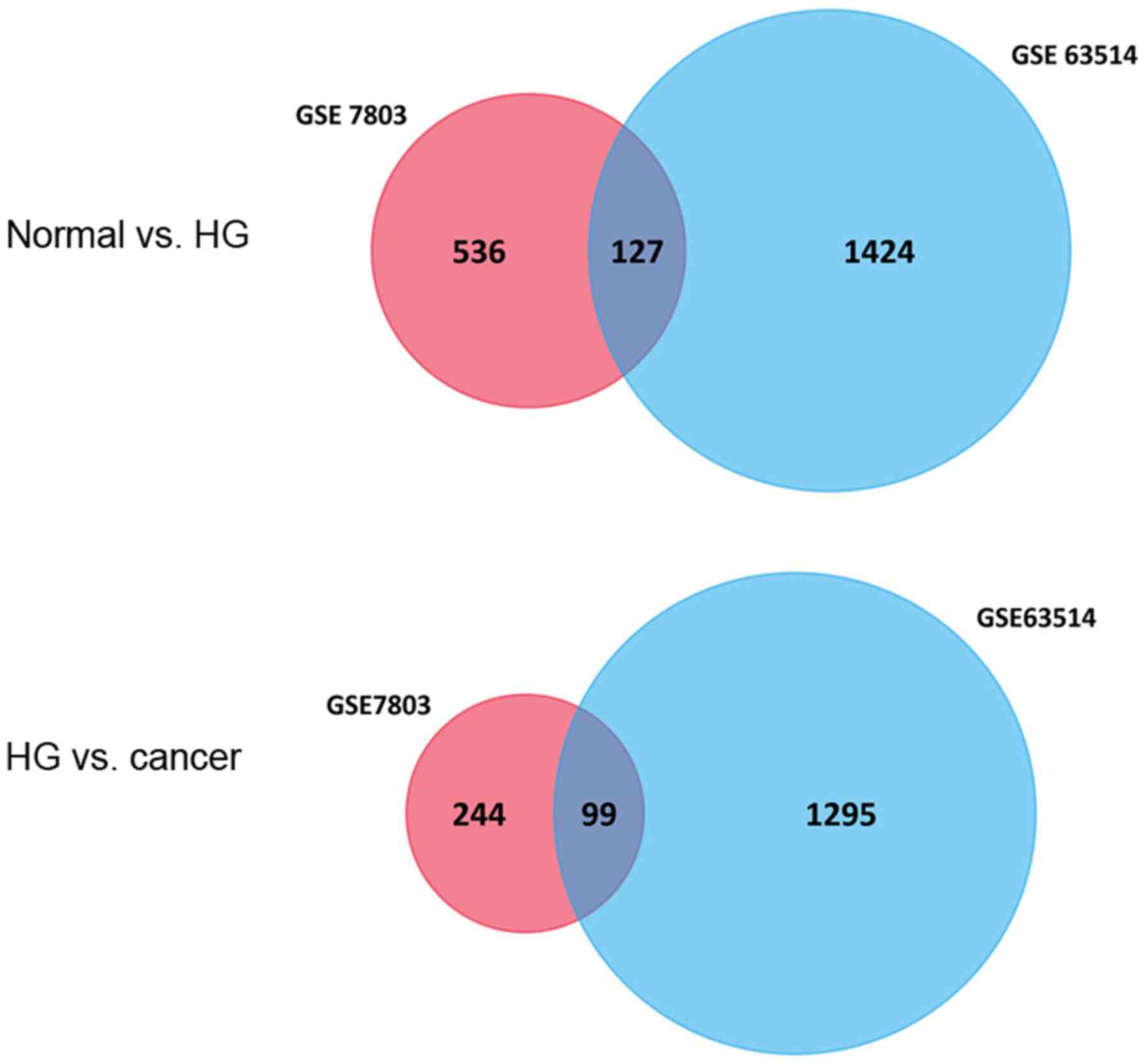
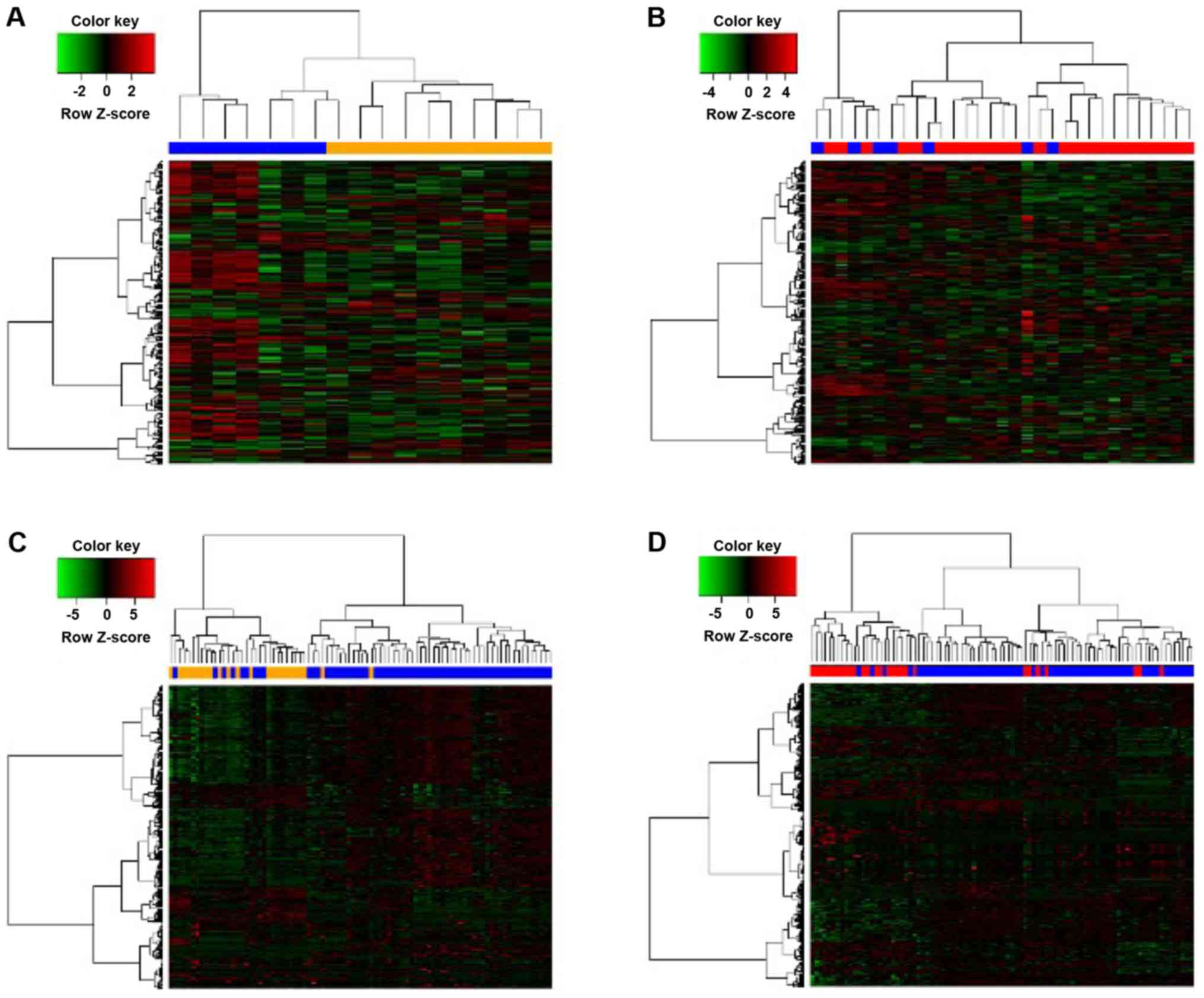
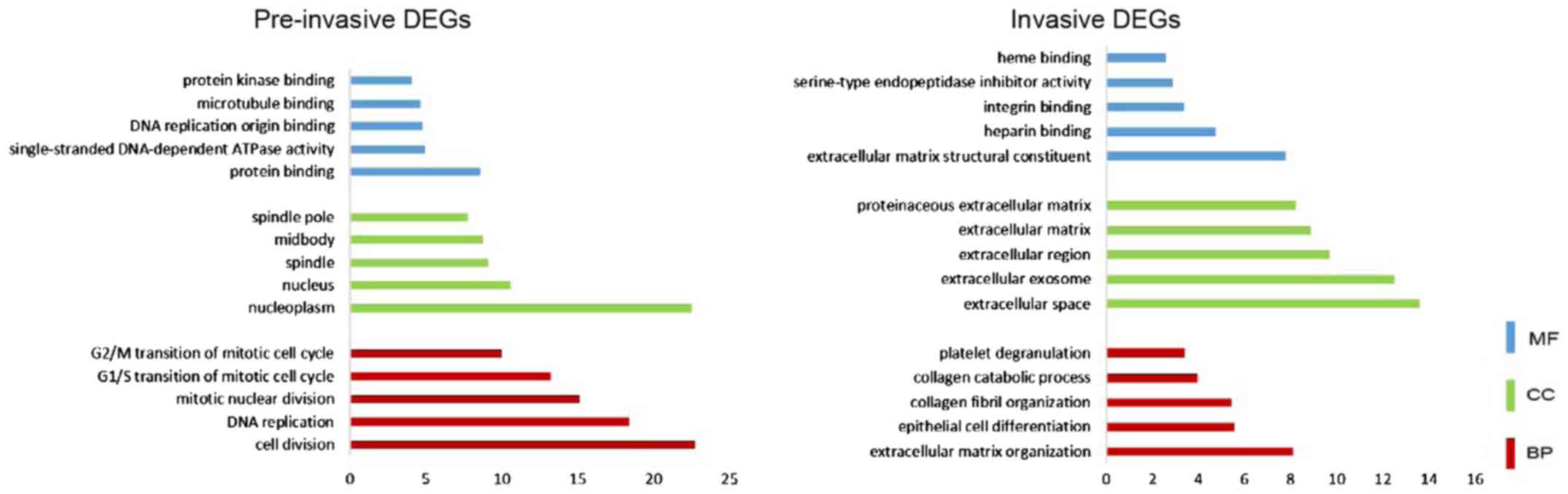
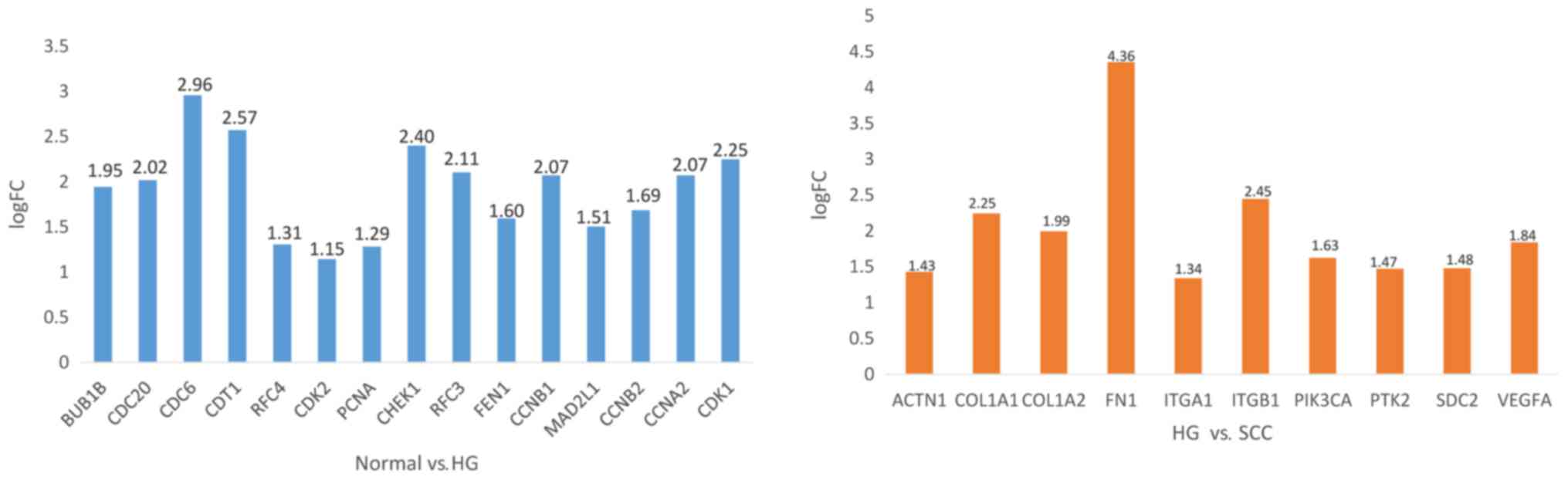
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vogelstein B and Kinzler KW: Cancer genes
and the pathways they control. Nat Med. 10:789–799. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu NL, Huang DY, Tsou HN, Lin YC and Lin
WW: Syk mediates IL-17-induced CCL20 expression by targeting
Act1-dependent k63-linked ubiquitination of TRAF6. J Invest
Dermatol. 135:490–498. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kulasingam V and Diamandis EP: Strategies
for discovering novel cancer biomarkers through utilization of
emerging technologies. Nat Clin Pract Oncol. 5:588–599. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu SF, Qian WY, Zhang JW, Yang YB, Liu Y,
Dong Y, Zhang ZB, Zhu YP and Feng YJ: Network motifs in the
transcriptional regulation network of cervical carcinoma cells
respond to EGF. Arch Gynecol Obstet. 287:771–777. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Luo Y, Wu Y, Peng Y, Liu X, Bie J and Li
S: Systematic analysis to identify a key role of CDK1 in mediating
gene interaction networks in cervical cancer development. Ir J Med
Sci. 185:231–239. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiao X, Sherman BT, Huang da W, Stephens
R, Baseler MW, Lane HC and Lempicki RA: DAVID-WS: A stateful web
service to facilitate gene/protein list analysis. Bioinformatics.
28:1805–1806. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gene Ontology Consortium: The Gene
Ontology (GO) project in 2006. Nucleic Acids Res. 34:(Database
Issue):. D322–D326. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: CytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8(Suppl 4): S112014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng J, Lu X, Wang J, Zhang H, Duan P and
Li C: Interactome analysis of gene expression profiles of cervical
cancer reveals dysregulated mitotic gene clusters. Am J Transl Res.
9:3048–3059. 2017.PubMed/NCBI
|
|
13
|
Cahill DP, Lengauer C, Yu J, Riggins GJ,
Willson JK, Markowitz SD, Kinzler KW and Vogelstein B: Mutations of
mitotic checkpoint genes in human cancers. Nature. 392:300–303.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ru HY, Chen RL, Lu WC and Chen JH: Hbub1
defects in leukemia and lymphoma cells. Oncogene. 21:4673–4679.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim Y, Choi JW, Lee JH and Kim YS: MAD2
and CDC20 are upregulated in high-grade squamous intraepithelial
lesions and squamous cell carcinomas of the uterine cervix. Int J
Gynecol Pathol. 33:517–523. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Masuda Y, Suzuki M, Piao J, Gu Y,
Tsurimoto T and Kamiya K: Dynamics of human replication factors in
the elongation phase of DNA replication. Nucleic Acids Res.
35:6904–6916. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lockwood WW, Thu KL, Lin L, Pikor LA,
Chari R, Lam WL and Beer DG: Integrative genomics identified RFC3
as an amplified candidate oncogene in esophageal adenocarcinoma.
Clin Cancer Res. 18:1936–1946. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Arai M, Kondoh N, Imazeki N, Hada A,
Hatsuse K, Matsubara O and Yamamoto M: The knockdown of endogenous
replication factor C4 decreases the growth and enhances the
chemosensitivity of hepatocellular carcinoma cells. Liver Int.
29:55–62. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Perez R, Wu N, Klipfel AA and Beart RW Jr:
A better cell cycle target for gene therapy of colorectal cancer:
Cyclin G. J Gastrointest Surg. 7:884–889. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
van Nimwegen MJ and van de Water B: Focal
adhesion kinase: A potential target in cancer therapy. Biochem
Pharmacol. 73:597–609. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Oktay MH, Oktay K, Hamele-Bena D, Buyuk A
and Koss LG: Focal adhesion kinase as a marker of malignant
phenotype in breast and cervical carcinomas. Hum Pathol.
34:240–245. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fong YC, Liu SC, Huang CY, Li TM, Hsu SF,
Kao ST, Tsai FJ, Chen WC, Chen CY and Tang CH: Osteopontin
increases lung cancer cells migration via activation of the
alphavbeta3 integrin/FAK/Akt and NF-kappaB-dependent pathway. Lung
Cancer. 64:263–270. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Moon HS, Park WI, Choi EA, Chung HW and
Kim SC: The expression and tyrosine phosphorylation of
E-cadherin/catenin adhesion complex, and focal adhesion kinase in
invasive cervical carcinomas. Int J Gynecol Cancer. 13:640–646.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Srinivasan M and Jewell SD: Quantitative
estimation of PCNA, c-myc, EGFR and TGF-alpha in oral submucous
fibrosis-an immunohistochemical study. Oral Oncol. 37:461–467.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lv Q, Zhang J, Yi Y, Huang Y, Wang Y, Wang
Y and Zhang W: Proliferating cell nuclear antigen has an
association with prognosis and risks factors of cancer patients: A
systematic review. Mol Neurobiol. 53:6209–6217. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim TH, Han JH, Shin E, Noh JH, Kim HS and
Song YS: Clinical implication of p16, Ki-67, and proliferating cell
nuclear antigen expression in cervical neoplasia: Improvement of
diagnostic accuracy for high-grade squamous intraepithelial lesion
and prediction of resection margin involvement on conization
specime. J Cancer Prev. 20:70–77. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gkiozos I, Tsagouli S, Charpidou A, Grapsa
D, Kainis E, Gratziou C and Syrigos K: Levels of vascular
endothelial growth factor in serum and pleural fluid are
independent predictors of survival in advanced non-small cell lung
cancer: Results of a prospective study. Anticancer Res.
35:1129–1137. 2015.PubMed/NCBI
|
|
28
|
Zhang J, Liu J, Zhu C, He J, Chen J, Liang
Y, Yang F, Wu X and Ma X: Prognostic role of vascular endothelial
growth factor in cervical cancer: A meta-analysis. Oncotarget.
8:24797–24803. 2017.PubMed/NCBI
|
|
29
|
Tsai HL, Yang IP, Lin CH, Chai CY, Huang
YH, Chen CF, Hou MF, Kuo CH, Juo SH and Wang JY: Predictive value
of vascular endothelial growth factor overexpression in early
relapse of colorectal cancer patients after curative resection. Int
J Colorectal Dis. 28:415–424. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tian RQ, Wang XH, Hou LJ, Jia WH, Yang Q,
Li YX, Liu M, Li X and Tang H: MicroRNA-372 is down-regulated and
targets cyclin-dependent kinase 2 (CDK2) and cyclin A1 in human
cervical cancer, which may contribute to tumorigene. J Biol Chem.
286:25556–25563. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Koncar RF, Feldman R, Bahassi EM and
Hashemi Sadraei N: Comparative molecular profiling of HPV-induced
squamous cell carcinomas. Cancer Med. 6:1673–1685. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
McIntyre JB, Wu JS, Craighead PS, Phan T,
Köbel M, Lees-Miller SP, Ghatage P, Magliocco AM and Doll CM:
PIK3CA mutational status and overall survival in patients with
cervical cancer treated with radical chemoradiotherapy. Gynecol
Oncol. 128:409–414. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chung TK, van Hummelen P, Chan PK, Cheung
TH, Yim SF, Yu MY, Ducar MD, Thorner AR, MacConaill LE, Doran G, et
al: Genomic aberrations in cervical adenocarcinomas in Hong Kong
Chinese women. Int J Cancer. 137:776–783. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cui B, Zheng B, Zhang X, Stendahl U,
Andersson S and Wallin KL: Mutation of PIK3CA: Possible risk factor
for cervical carcinogenesis in older women. Int J Oncol.
34:409–416. 2009.PubMed/NCBI
|