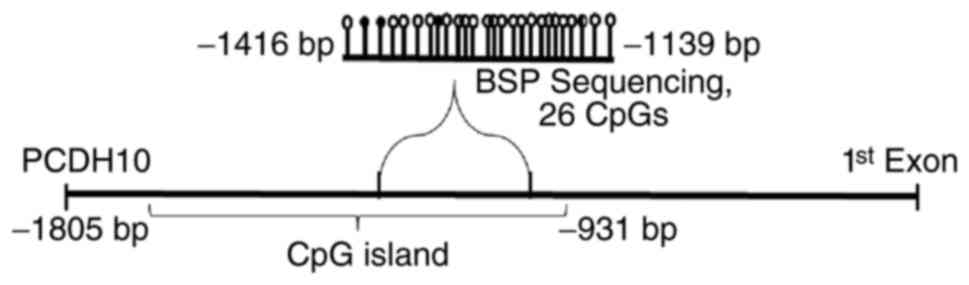
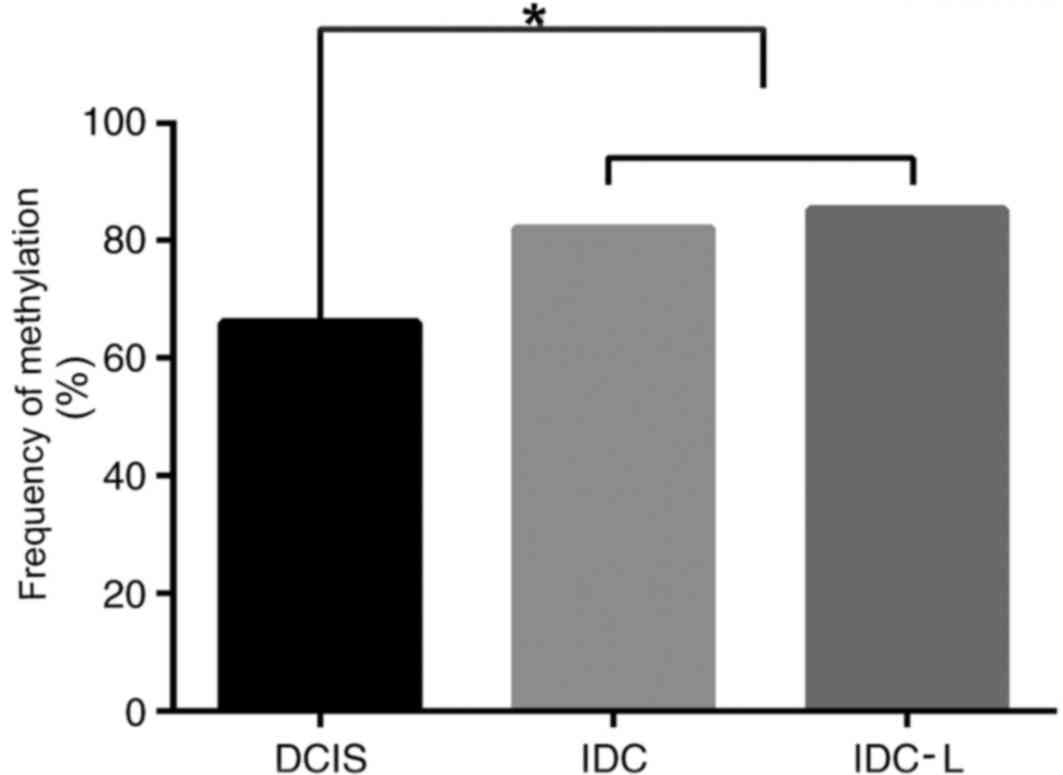
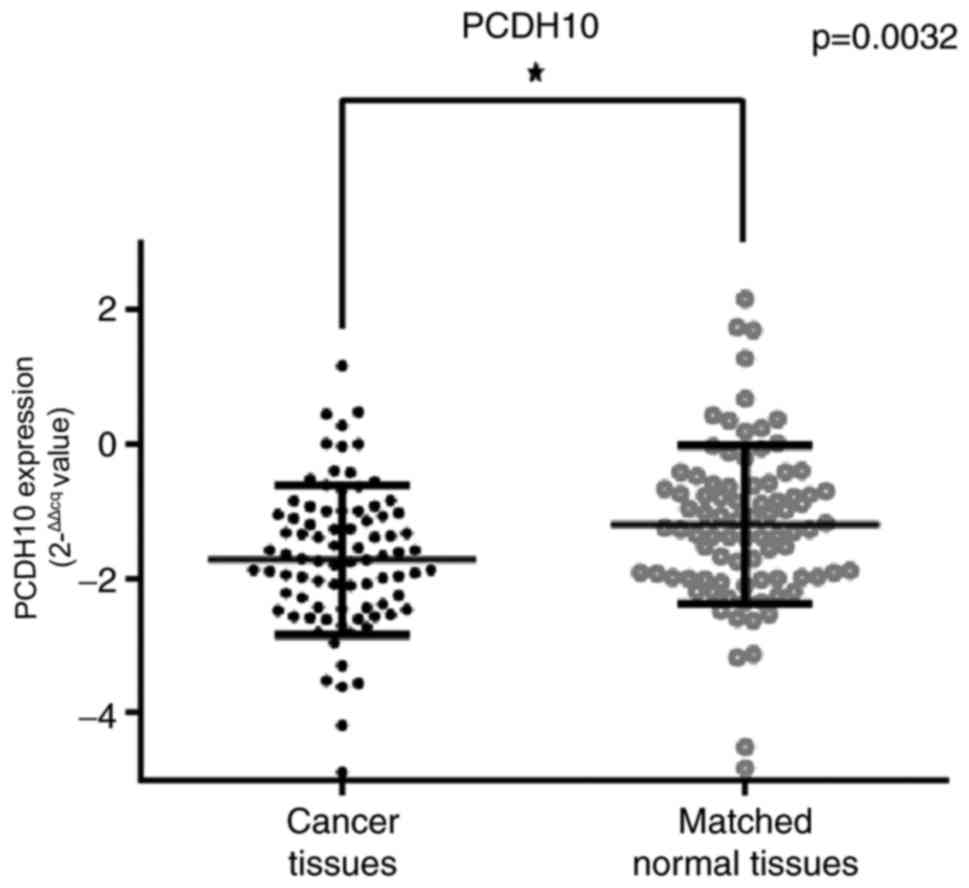
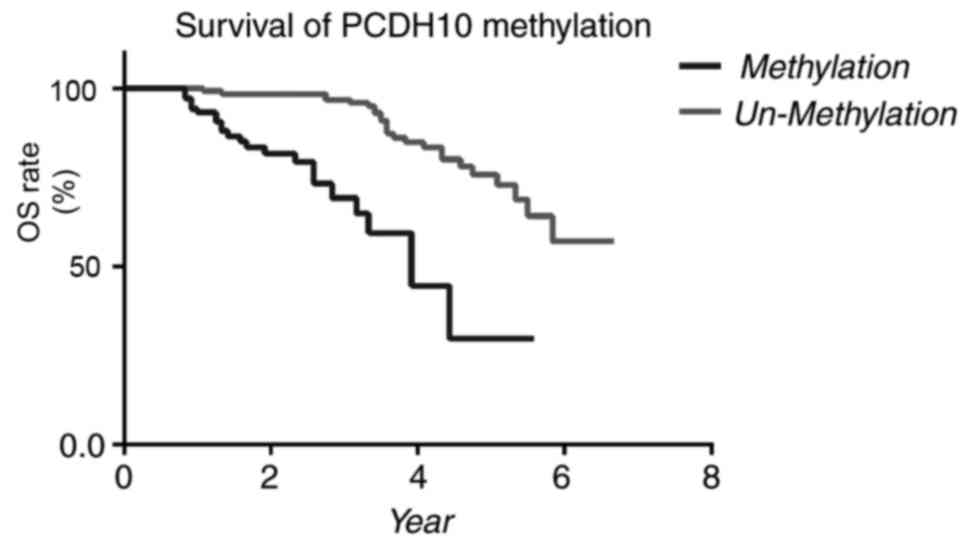
|
1
|
Seneviratne S, Lawrenson R, Scott N, Kim
B, Shirley R and Campbell I: Breast cancer biology and ethnic
disparities in breast cancer mortality in new zealand: A cohort
study. PLoS One. 10:e01235232015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast
cancer in China. Lancet Oncol. 15:e279–89. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Frazer KA, Murray SS, Schork NJ and Topol
EJ: Human genetic variation and its contribution to complex traits.
Nat Rev Genet. 10:241–251. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rivera CM and Ren B: Mapping human
epigenomes. Cell. 155:39–55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maurano MT, Humbert R, Rynes E, Thurman
RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et
al: Systematic localization of common disease-associated variation
in regulatory DNA. Science. 337:1190–1195. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baylin SB and Ohm JE: Epigenetic gene
silencing in cancer-a mechanism for early oncogenic pathway
addiction? Nat Rev Cancer. 6:107–116. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Feinberg AP, Ohlsson R and Henikoff S: The
epigenetic progenitor origin of human cancer. Nat Rev Genet.
7:21–33. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Esteller M, Fraga MF, Guo M,
Garcia-Foncillas J, Hedenfalk I, Godwin AK, Trojan J,
Vaurs-Barrière C, Bignon YJ, Ramus S, et al: DNA methylation
patterns in hereditary human cancers mimic sporadic tumorigenesis.
Hum Mol Genet. 10:3001–3007. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Craig JM and Bickmore WA: The distribution
of CpG islands in mammalian chromosomes. Nat Genet. 7:376–382.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Eden A, Gaudet F, Waghmare A and Jaenisch
R: Chromosomal instability and tumors promoted by DNA
hypomethylation. Science. 300:4552003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Esteller M: Epigenetics in cancer. N Engl
J Med. 358:1148–1159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fernandez AF, Assenov Y, Martin-Subero JI,
Balint B, Siebert R, Taniguchi H, Yamamoto H, Hidalgo M, Tan AC,
Galm O, et al: A DNA methylation fingerprint of 1628 human samples.
Genome Res. 22:407–419. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moarii M, Boeva V, Vert JP and Reyal F:
Changes in correlation between promoter methylation and gene
expression in cancer. BMC Genomics. 16:8732015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu YY, Sun CX, Liu YK, Li Y, Wang L and
Zhang W: Genome-wide screen of ovary-specific DNA methylation in
polycystic ovary syndrome. Fertil Steril. 104(145–153): e62015.
|
|
16
|
Esteller M: Cancer epigenomics: DNA
methylomes and histone-modification maps. Nat Rev Genet. 8:286–298.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Van Neste L, Herman JG, Otto G, Bigley JW,
Epstein JI and Van Criekinge W: The epigenetic promise for prostate
cancer diagnosis. Prostate. 72:1248–1261. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lo PK and Sukumar S: Epigenomics and
breast cancer. Pharmacogenomics. 9:1879–1902. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shan M, Su Y, Kang W, Gao R, Li X and
Zhang G: Aberrant expression and functions of protocadherins in
human malignant tumors. Tumour Biol. 37:12969–12981. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Otani K, Li X, Arakawa T, Chan FK and Yu
J: Epigenetic-mediated tumor suppressor genes as diagnostic or
prognostic biomarkers in gastric cancer. Expert Rev Mol Diagn.
13:445–455. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu J, Cheng YY, Tao Q, Cheung KF, Lam CN,
Geng H, Tian LW, Wong YP, Tong JH, Ying JM, et al: Methylation of
protocadherin 10, a novel tumor suppressor, is associated with poor
prognosis in patients with gastric cancer. Gastroenterology.
136(640–651): e12009.
|
|
22
|
Lv J, Zhu P, Yang Z, Li M, Zhang X, Cheng
J, Chen X and Lu F: PCDH20 functions as a tumour-suppressor gene
through antagonizing the Wnt/β-catenin signalling pathway in
hepatocellular carcinoma. J Viral Hepat. 22:201–211. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang C, Peng Y, Yang F, Qin R, Liu W and
Zhang C: PCDH8 is frequently inactivated by promoter
hypermethylation in liver cancer: Diagnostic and clinical
significance. J Cancer. 7:446–452. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee NK, Lee JH, Kim WK, Yun S, Youn YH,
Park CH, Choi YY, Kim H and Lee SK: Promoter methylation of PCDH10
by HOTAIR regulates the progression of gastrointestinal stromal
tumors. Oncotarget. 7:75307–75318. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miyamoto K, Fukutomi T, Akashi-Tanaka S,
Hasegawa T, Asahara T, Sugimura T and Ushijima T: Identification of
20 genes aberrantly methylated in human breast cancers. Int J
Cancer. 116:407–414. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ying J, Li H, Seng TJ, Langford C,
Srivastava G, Tsao SW, Putti T, Murray P, Chan AT and Tao Q:
Functional epigenetics identifies a protocadherin PCDH10 as a
candidate tumor suppressor for nasopharyngeal, esophageal and
multiple other carcinomas with frequent methylation. Oncogene.
25:1070–1080. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Foulkes WD: Inherited susceptibility to
common cancers. N Engl J Med. 359:2143–2153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pharoah PD, Day NE, Duffy S, Easton DF and
Ponder BA: Family history and the risk of breast cancer: A
systematic review and meta-analysis. Int J Cancer. 71:800–809.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Porter P: ‘Westernizing’ women's risks?
Breast cancer in lower-income countries. N Engl J Med. 358:213–216.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lynch HT, Silva E, Snyder C and Lynch JF:
Hereditary breast cancer: Part I. Diagnosing hereditary breast
cancer syndromes. Breast J. 14:3–13. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Majdak-Paredes EJ and Fatah F: Hereditary
breast cancer syndromes and clinical implications. J Plast Reconstr
Aesthet Surg. 62:181–189. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Harvey JM, Clark GM, Osborne CK and Allred
DC: Estrogen receptor status by immunohistochemistry is superior to
the ligand-binding assay for predicting response to adjuvant
endocrine therapy in breast cancer. J Clin Oncol. 17:1474–1481.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ogura H, Akiyama F, Kasumi F, Kazui T and
Sakamoto G: Evaluation of HER-2 status in breast carcinoma by
fluorescence in situ hybridization and immunohistochemistry. Breast
Cancer. 10:234–240. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Soong R, Robbins PD, Dix BR, Grieu F, Lim
B, Knowles S, Williams KE, Turbett GR, House AK and Iacopetta BJ:
Concordance between p53 protein overexpression and gene mutation in
a large series of common human carcinomas. Hum Pathol.
27:1050–1055. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hugh J, Hanson J, Cheang MC, Nielsen TO,
Perou CM, Dumontet C, Reed J, Krajewska M, Treilleux I, Rupin M, et
al: Breast cancer subtypes and response to docetaxel in
node-positive breast cancer: Use of an immunohistochemical
definition in the BCIRG 001 trial. J Clin Oncol. 27:1168–1176.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu Y, Yan W, Liu X, Jia Y, Cao B, Yu Y, Lv
Y, Brock MV, Herman JG, Licchesi J, et al: DACT2 is frequently
methylated in human gastric cancer and methylation of DACT2
activated Wnt signaling. Am J Cancer Res. 4:710–724.
2014.PubMed/NCBI
|
|
37
|
Koontz L: Agarose gel electrophoresis.
Methods Enzymol. 529:35–45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Eads CA, Danenberg KD, Kawakami K, Saltz
LB, Blake C, Shibata D, Danenberg PV and Laird PW: MethyLight: A
high-throughput assay to measure DNA methylation. Nucleic Acids
Res. 28:E322000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kagan J, Srivastava S, Barker PE, Belinsky
SA and Cairns P: Towards clinical application of methylated DNA
sequences as cancer biomarkers: A Joint NCI's EDRN and NIST
workshop on standards, methods, assays, reagents and tools. Cancer
Res. 67:4545–4549. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ogino S, Kawasaki T, Brahmandam M, Cantor
M, Kirkner GJ, Spiegelman D, Makrigiorgos GM, Weisenberger DJ,
Laird PW, Loda M and Fuchs CS: Precision and performance
characteristics of bisulfite conversion and real-time PCR
(MethyLight) for quantitative DNA methylation analysis. J Mol
Diagn. 8:209–217. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Widschwendter M, Siegmund KD, Müller HM,
Fiegl H, Marth C, Müller-Holzner E, Jones PA and Laird PW:
Association of breast cancer DNA methylation profiles with hormone
receptor status and response to tamoxifen. Cancer Res.
64:3807–3813. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Widschwendter M, Apostolidou S, Jones AA,
Fourkala EO, Arora R, Pearce CL, Frasco MA, Ayhan A, Zikan M,
Cibula D, et al: HOXA methylation in normal endometrium from
premenopausal women is associated with the presence of ovarian
cancer: A proof of principle study. Int J Cancer. 125:2214–2218.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cho YH, Shen J, Gammon MD, Zhang YJ, Wang
Q, Gonzalez K, Xu X, Bradshaw PT, Teitelbaum SL, Garbowski G, et
al: Prognostic significance of gene-specific promoter
hypermethylation in breast cancer patients. Breast Cancer Res
Treat. 131:197–205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang W and Srivastava S: Strategic
approach to validating methylated genes as biomarkers for breast
cancer. Cancer Prev Res (Phila). 3:16–24. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Clark SJ and Melki J: DNA methylation and
gene silencing in cancer: Which is the guilty party? Oncogene.
21:5380–5387. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Muggerud AA, Rønneberg JA, Wärnberg F,
Botling J, Busato F, Jovanovic J, Solvang H, Bukholm I,
Børresen-Dale AL, Kristensen VN, et al: Frequent aberrant DNA
methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma
in situ and early invasive breast cancer. Breast Cancer Res.
12:R32010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wolverton T and Lalande M: Identification
and characterization of three members of a novel subclass of
protocadherins. Genomics. 76:66–72. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Qiu GH, Tan LK, Loh KS, Lim CY, Srivastava
G, Tsai ST, Tsao SW and Tao Q: The candidate tumor suppressor gene
BLU, located at the commonly deleted region 3p21.3, is an
E2F-regulated, stress-responsive gene and inactivated by both
epigenetic and genetic mechanisms in nasopharyngeal carcinoma.
Oncogene. 23:4793–4806. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jeschke J, Van Neste L, Glöckner SC, Dhir
M, Calmon MF, Deregowski V, Van Criekinge W, Vlassenbroeck I, Koch
A, Chan TA, et al: Biomarkers for detection and prognosis of breast
cancer identified by a functional hypermethylome screen.
Epigenetics. 7:701–709. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Morris MR, Ricketts C, Gentle D,
Abdulrahman M, Clarke N, Brown M, Kishida T, Yao M, Latif F and
Maher ER: Identification of candidate tumour suppressor genes
frequently methylated in renal cell carcinoma. Oncogene.
29:2104–2117. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mendonsa AM, Na TY and Gumbiner BM:
E-cadherin in contact inhibition and cancer. Oncogene. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Su YJ, Chang YW, Lin WH, Liang CL and Lee
JL: An aberrant nuclear localization of E-cadherin is a potent
inhibitor of Wnt/beta-catenin-elicited promotion of the cancer stem
cell phenotype. Oncogenesis. 4:e1572015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Patra SK and Bettuzzi S: Epigenetic
DNA-methylation regulation of genes coding for lipid
raft-associated components: A role for raft proteins in cell
transformation and cancer progression (review). Oncol Rep.
17:1279–1290. 2007.PubMed/NCBI
|
|
55
|
Imoto I, Izumi H, Yokoi S, Hosoda H,
Shibata T, Hosoda F, Ohki M, Hirohashi S and Inazawa J: Frequent
silencing of the candidate tumor suppressor PCDH20 by epigenetic
mechanism in non-small-cell lung cancers. Cancer Res. 66:4617–4626.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Haruki S, Imoto I, Kozaki K, Matsui T,
Kawachi H, Komatsu S, Muramatsu T, Shimada Y, Kawano T and Inazawa
J: Frequent silencing of protocadherin 17, a candidate tumour
suppressor for esophageal squamous cell carcinoma. Carcinogenesis.
31:1027–1036. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Uyen TN, Sakashita K, Al-Kzayer LF,
Nakazawa Y, Kurata T and Koike K: Aberrant methylation of
protocadherin 17 and its prognostic value in pediatric acute
lymphoblastic leukemia. Pediatr Blood Cancer. 64:2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Qiu C, Bu X and Jiang Z: Protocadherin-10
acts as a tumor suppressor gene, and is frequently downregulated by
promoter methylation in pancreatic cancer cells. Oncol Rep.
36:383–389. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shi D, Murty VV and Gu W: PCDH10, a novel
p53 transcriptional target in regulating cell migration. Cell
Cycle. 14:857–866. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yang Y, Jiang Y, Jiang M, Zhang J, Yang B,
She Y, Wang W, Deng Y and Ye Y: Protocadherin 10 inhibits cell
proliferation and induces apoptosis via regulation of DEP domain
containing 1 in endometrial endometrioid carcinoma. Exp Mol Pathol.
100:344–352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang KH, Liu HW, Lin SR, Ding DC and Chu
TY: Field methylation silencing of the protocadherin 10 gene in
cervical carcinogenesis as a potential specific diagnostic test
from cervical scrapings. Cancer Sci. 100:2175–2180. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Tang X, Yin X, Xiang T, Li H, Li F, Chen L
and Ren G: Protocadherin 10 is frequently downregulated by promoter
methylation and functions as a tumor suppressor gene in non-small
cell lung cancer. Cancer Biomark. 12:11–19. 2013. View Article : Google Scholar
|
|
63
|
Li Y, Yang ZS, Song JJ, Liu Q and Chen JB:
Protocadherin-10 is involved in angiogenesis and methylation
correlated with multiple myeloma. Int J Mol Med. 29:704–710. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhong X, Zhu Y, Mao J, Zhang J and Zheng
S: Frequent epigenetic silencing of PCDH10 by methylation in human
colorectal cancer. J Cancer Res Clin Oncol. 139:485–490. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Novak P, Jensen TJ, Garbe JC, Stampfer MR
and Futscher BW: Stepwise DNA methylation changes are linked to
escape from defined proliferation barriers and mammary epithelial
cell immortalization. Cancer Res. 69:5251–5258. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Pimson C, Ekalaksananan T, Pientong C,
Promthet S, Putthanachote N, Suwanrungruang K and Wiangnon S:
Aberrant methylation of PCDH10 and RASSF1A genes in blood samples
for non-invasive diagnosis and prognostic assessment of gastric
cancer. Peer J. 4:e21122016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hou YC, Deng JY, Zhang RP, Xie XM, Cui JL,
Wu WP, Hao XS and Liang H: Evaluating the clinical feasibility: The
direct bisulfite genomic sequencing for examination of methylated
status of protocadherin10 (PCDH10) promoter to predict the
prognosis of gastric cancer. Cancer Biomark. 15:567–573. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Deng QK, Lei YG, Lin YL, Ma JG and Li WP:
Prognostic value of Protocadherin10 (PCDH10) methylation in serum
of prostate cancer patients. Med Sci Monit. 22:516–521. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Donkin I and Barres R: Sperm epigenetics
and influence of environmental factors. Mol Metab. Feb
27–2018.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ingerslev LR, Donkin I, Fabre O, Versteyhe
S, Mechta M, Pattamaprapanont P, Mortensen B, Krarup NT and Barrès
R: Endurance training remodels sperm-borne small RNA expression and
methylation at neurological gene hotspots. Clin Epigenetics.
10:122018. View Article : Google Scholar : PubMed/NCBI
|