Introduction
It has been reported that the female cervix
maintains communities of microbial species, which have a symbiotic
relationship with the host (1). It
has been demonstrated that the female cervix is colonized by
diverse microbiota, which serve a crucial role in cervicovaginal
health (1). However, it has been
indicated that the type of organisms present is dependent on the
prevailing environmental conditions and host factors (1,2). Over the
last decade, it has been indicated that the development and
introduction of molecular-based technology has provided novel
information regarding the composition of the vaginal-cervical
flora, as well as the abnormal colonization of the genital tract by
pathogens (2). The aforementioned
findings may help to elucidate the microbiome of the genital tract
in healthy women and in human papilloma virus (HPV)-dependent
carcinogenesis.
The majority of previous studies have indicated that
the cervical/vaginal microbial flora has a prevalence of
Lactobacillus species, which produce lactic acids that
maintain an acidic environment and may inhibit pathogenic growth
(2–8).
Specifically, lactic acid and other related acidic compounds have
been reported to inhibit bacterial growth associated with bacterial
vaginosis (BV), as well as viral infections (3). In addition, lactic acid has been
recognized as a component of the immune defence system, as it has
been demonstrated to potentiate the production of protective
proinflammatory cytokines by vaginal epithelial cells, to promote
the activation of T helper 17 lymphocytes, to stimulate dendritic
cell maturation and induce interferon production (1). Over 120 Lactobacillus species
have been identified and over 20 species have been identified in
the vagina (2).
The dominant microorganisms in the vagina of a
healthy woman during puberty have been reported to be from the
Lactobacillus genus, including L. acidophilus, L.
fermentum, L. plantarum, L. brevis, L. jenseni, L. casei, L.
catenaforme, L. delbrueckii and L. salivarius (1,2). It is
considered that in the majority of cases, vaginal inflammation is
not caused by novel microorganisms introduced from the outside, but
instead by a disturbance to the proportion and number of
microorganisms already existing in the vagina (4). Each bacterium has been reported as a
potential etiological factor of inflammation, including
Lactobacillus (4). It has been
reported that anaerobic bacteria have a significantly increased
pathogenic potential compared with aerobic bacteria (5,6). Factors,
which have been reported to potentially disturb vaginal biocenosis
include the following hormonal changes: Pregnancy, puberty,
menopause and hormonal contraception, particularly using low doses
of estrogen; vaginal sterilization following chemo- or antibiotic
therapy; surgical conditions such as vaginoplasty, erosions, poor
genitalia hygiene, including the vulva and the vagina, and lack of
a regimented sex life, i.e. frequent, unprotected, lack of a
regular sex life (4–6). Large variation in the number and
composition of bacteria has been reported among women, and among
time intervals for one woman (5,6).
Therefore, attempting to evaluate the vaginal microbiome is
extremely difficult.
A healthy cervicovaginal microenvironment has been
reported to be characterised by high levels of different species of
Lactobacillus, including the predominant L. crispatus, L.
iners, L. jensenii and L. gasseri. Other species may
occur occasionally (2,6). In rare cases, it has been reported that
the cervix can be colonized by the same two or four
Lactobacillus species (4). The
aforementioned cases have been demonstrated to be dependent on a
number of genetic and environmental factors, including nationality,
diet and age (2,6). A number of previous studies have
indicated that, in a significant proportion of healthy women, the
Lactobacillus in the vagina may be replaced by other lactic
acid-producing bacteria, including Atopobium vaginae,
Megaspharea and Leptotrichia species (3). It has been indicated that an abnormal
vaginal microenvironment may be caused by sexually transmitted
infection (3). It has been reported
that trichomoniasis may be caused by colonization with a
microorganism not commonly identified within vaginal colonies,
including Streptococcus pneumoniae, Haemophilus influenzae
and Listeria monocytogenes (6,7). In
addition, it has been reported that an abnormal microenvironment
may be caused by an invasion of an alternative organism, which is a
component of the normal vaginal flora, including Escherichia
coli (3). Bacterial vaginosis has
been reported as a disorder characterised by a decrease in the
quality or quantity of Lactobacillus and the growth of
Mycoplasma hominis, Gardnerella vaginalis, Mobiluncus
species, Neisseria gonorrhea, Trichomonas vaginalis, Chlamydia
trachomatis and Prevotella species (4,5). A
previous meta-analysis reported a positive association between
cervical HPV infection and BV (6,7). The
reverse phenomenon of HPV as a risk factor for BV invasion has also
been described (8).
It has been demonstrated that HPV is considered a
principal factor responsible for the development of cervical cancer
(8). However, it has been suggested
that HPV infection alone does not cause cervical carcinogenesis and
that other factors, such as prolonged oral contraceptive or
smoking, may be involved in the disease progression (8). To determine whether the vaginal
microflora is affected by one of these factors, the present study
investigated the association between cervical and vaginal
infections, and pre-cancerous lesions of the cervix.
Materials and methods
Patient samples
Women included in the present study were recruited
from cervical cancer screening in the Department of Gynaecological
Oncology and Gynaecology, Medical University of Lublin in Poland
between February 2003 and August 2015. The study group consisted of
250 women. Patients whose molecular analysis indicated that they
were HPV-positive [HPV(+)] were included within the analysed group
(n=180), whereas the healthy women [HPV(−)] were included within
the control group (n=70).
Cytology and HPV status were used to classify the
patients into the following 3 groups: Control group [n=70);
HPV(−)]; women with low-grade squamous intraepithelial lesions
[LSIL; n=95; HPV(−)], and women with high-grade squamous
intraepithelial lesions [HSIL; n=85; HPV(+)]. The mean ages of
patients with LSIL and HSIL, and the control group, did not
significantly differ: 35 years (range, 25–48) and 37 years (range,
29–48) compared with 37 years (range 21–48), respectively
(P>0.05). The present study was approved by the Ethics Committee
of the Medical University of Lublin (Lublin, Poland). Written
informed consent was obtained from all participants in the present
study and the research was performed in accordance with the
Declaration of Helsinki.
The patient inclusion criteria for the present study
for the research and control groups were as follows: i) Cytological
diagnosis of LSIL or HSIL or patients with a normal cytological
swab; ii) HPV (+) or (−) status; iii) no use of oral and/or vaginal
probiotics for 30 days prior to the start of the present study; iv)
absence of genital tract infection during the 30 days prior to the
classification of patients in the study, and v) a maximum of one
sexual partner for 30 days prior to qualification testing.
The exclusion criteria for participating in the
present study included the following: i) Vaginal bleeding of
unknown etiology; ii) pregnancy; iii) oral contraceptive use or
hormone replacement therapy; iv) cigarette smoking; v) history of
other types of cancer; vi) systemic diseases; vii) diabetes, and
viii) thyroid or other endocrine diseases.
Cervical swabs were collected from patients and to
rule out experimental bias or random error. pooling was performed
in 3 subgroups as follows: in the control group 23, 23, 24 swabs
were pooled; in the LSIL group 31, 32, 32 were pooled, and in the
HSIL group 28, 28, 29 swabs were pooled. The pooled cervical swabs
were stored immediately at −80°C for a maximum 12 months. The
cervical cytological findings were classified according to the
Bethesda system (9).
DNA isolation from the swabs
Total DNA was extracted from cells using a DNA
isolation kit (QIAamp DNA kit; cat. no. 51306; Qiagen GmbH, Hilden,
Germany), according to the manufacturer's protocols.
Identification of HPV DNA
Identification of HPV-derived DNA was performed by
polymerase chain reaction (PCR) amplification of HPV gene
sequences. The MY09, MY11, LC1 and LC2 primers (Institute of
Biochemistry and Biophysics Polish Academy of Sciences, Warsaw,
Poland) that were complementary to the genomic sequence of the
predominantly diagnosed HPV types were used, as previously
described (10).
Typing of 16S ribosomal DNA (16S rRNA)
by PCR-amplification and next generation sequencing
The V4 hypervariable regions of 16S rRNA in
bacterial genes were detected by PCR, as previously described
(11). Sequencing and sequence
analysis was performed at Genomed S.A, Warszawa (Poland).
Statistical analysis
The difference in the mean age of the female
patients was tested using the Kruskal-Wallis test and was
statistically significant if the calculated P>0.05. Using the
16S rRNA gene sequencing data, the frequency of bacteria
occurrences were calculated by multiplying the total number of
present bacteria with the percentage of bacterial concentration in
the cervical swabs. The statistical analysis was performed using
k-means cluster analysis (k-means clustering algorithm) and
Statistica 12.0 software (StatSoft Inc., Cracow, Poland).
Results
Identification and cluster analysis of
bacteria in the control, LSIL HPV(+) and HSIL HPV(+) groups
16S rRNA sequence-based methods were used to
identify healthy cervical microbial colonies, as well as those
associated with HPV-dependent carcinogenesis. The identified
microflora were grouped into CSTs as follows: Control, HPV(−);
LSIL, HPV(+), and HSIL HPV(+). A total of 6,466 bacteria species,
belonging to 74 classes, were identified from the vaginal/cervical
swabs of the patients. The major classes observed are presented in
Table I.
 | Table I.The frequency of bacteria classes in
vaginal/cervical swabs. |
Table I.
The frequency of bacteria classes in
vaginal/cervical swabs.
| Class | Healthy (%) | LSIL HPV(+)
(%) | HSIL HPV(+)
(%) |
|---|
|
Actinobacteria | 0.21 | 1.0 | 8.10 |
|
Alphaproteobacteria | 0.20 | 0.03 | 0.41 |
| Bacilli | 96.27 | 84.00 | 27.69 |
|
Bacterioidia | 0.01 | 0.01 | 0.00 |
|
Betaproteobacteria | 0.01 | 0.01 | 0.01 |
|
Clostridia | 0.09 | 0.16 | 0.20 |
|
Deltaproteobacteria | 0.00 | 0.00 | 0.01 |
|
Flavobacteriia | 0.01 | 0.00 | 0.03 |
|
Gammproteobacteria | 0.19 | 8.20 | 61.48 |
|
Ktedonobactetria | 0.01 | 0.00 | 0.00 |
|
Methanomicrobiota | 0.00 | 0.01 | 0.00 |
|
Mollicutes | 0.25 | 0.03 | 0.00 |
|
Nostocophycideae | 0.41 | 0.02 | 0.15 |
|
Sphingobacteria | 0.00 | 0.01 | 0.01 |
| Unclassified | 2.34–2.63 |
|
|
Identification and cluster analysis of
bacterial classes in the control, LSIL HPV(+) and HSIL HPV(+)
groups
Frequency analysis of bacterial classes in the CST
healthy women, as well as in patients with a diagnosis of LSIL
HPV(+) or HSIL HPV(+) revealed the presence of three major
clusters. The results of the bacterial classes cluster analysis are
presented in Table II. The bacterial
classes, which were distinct from the other classes regardless of
the type of diagnosis, included Bacilli, Actinobacteria and
Gammaproteobacteria (Table
II). Analysis of the frequencies of the individual classes
demonstrated that the CST composition in the healthy women, as well
as the women diagnosed with LSIL was formed predominantly of the
Bacilli bacteria. In CST patients with cancer, the
Gammaproteobacteria class was additionally detectable.
Further cluster analysis, from classes to species, was performed to
determine the bacterial species making up the three selected
bacteria classes.
 | Table II.Three major clusters of bacteria
classes in volunteers and patients with LSIL and HSIL
diagnosis. |
Table II.
Three major clusters of bacteria
classes in volunteers and patients with LSIL and HSIL
diagnosis.
| Control | LSIL HPV(+) | HSIL HPV(+) |
|---|
|
|
|
|---|
| Class | Gr. | Dist. | Class | Gr. | Dist. | Class | Gr. | Dist. |
|---|
| Bacilli | 1 | 0.00 | Bacilli | 1 | 0.00 | Bacilli | 1 | 1255.09 |
|
Actinobacteria | 2 | 3.01 |
Gammaproteobacteria | 1 | 0.00 |
Gammaproteobacteria | 1 | 1255.09 |
|
Alphaproteobacteria | 2 | 4.44 |
Actinobacteria | 2 | 25.94 |
Actinobacteria | 2 | 0.00 |
|
Gammaproteobacteria | 2 | 5.32 |
Bacteroidia | 2 | 33.90 |
Alphaproteobacteria | 3 | 23.48 |
|
Mollicutes | 2 | 0.38 |
Clostridia | 2 | 30.74 |
Bacteroidia | 3 | 2.23 |
|
Nostocophycideae | 2 | 13.16 |
Alphaproteobacteria | 3 | 20.47 |
Betaproteobacteria | 3 | 1.69 |
|
Bacteroidia | 3 | 0.14 |
Betaproteobacteria | 3 | 2.14 |
Chrysiogenetes | 3 | 2.48 |
|
Betaproteobacteria | 3 | 0.14 |
Chrysiogenetes | 3 | 2.82 |
Clostridia | 3 | 11.07 |
|
Chrysiogenetes | 3 | 0.50 |
Deferribacteres | 3 | 2.78 |
Deferribacteres | 3 | 2.53 |
|
Clostridia | 3 | 7.07 |
Deinococci | 3 | 2.82 |
Deinococci | 3 | 2.48 |
|
Deferribacteres | 3 | 0.50 |
Deltaproteobacteria | 3 | 2.75 |
Deltaproteobacteria | 3 | 2.06 |
|
Deinococci | 3 | 0.50 |
Epsilonproteobacteria | 3 | 3.25 |
Epsilonproteobacteria | 3 | 2.37 |
|
Deltaproteobacteria | 3 | 0.50 |
Erysipelotrichi | 3 | 2.83 |
Erysipelotrichi | 3 | 2.53 |
|
Epsilonproteobacteria | 3 | 0.42 |
Flavobacteriia | 3 | 2.26 |
Flavobacteriia | 3 | 1.58 |
|
Erysipelotrichi | 3 | 0.50 |
Fusobacteria | 3 | 2.68 |
Fusobacteria | 3 | 2.53 |
|
Fusobacteria | 3 | 0.34 |
Ktedonobacteria | 3 | 2.08 |
Ktedonobacteria | 3 | 2.37 |
|
Ktedonobacteria | 3 | 0.30 |
Methanomicrobia | 3 | 2.56 |
Methanomicrobia | 3 | 2.48 |
|
Methanomicrobia | 3 | 0.42 |
Mollicutes | 3 | 2.80 |
Mollicutes | 3 | 7.58 |
|
Opitutae | 3 | 0.50 |
Nostocophycideae | 3 | 33.15 |
Nostocophycideae | 3 | 11.59 |
|
Oscillatoriophycideae | 3 | 0.26 |
Opitutae | 3 | 2.82 |
Opitutae | 3 | 2.37 |
|
Planctomycetia | 3 | 0.50 |
Oscillatoriophycideae | 3 | 2.42 |
Oscillatoriophycideae | 3 | 2.32 |
|
Rubrobacteria | 3 | 0.50 |
Planctomycetia | 3 | 2.80 |
Planctomycetia | 3 | 2.53 |
|
Sphingobacteriia | 3 | 0.26 |
Rubrobacteria | 3 | 2.82 |
Rubrobacteria | 3 | 2.53 |
|
Synechococcophycideae | 3 | 0,50 |
Sphingobacteriia | 3 | 2.96 |
Sphingobacteriia | 3 | 1.43 |
|
Synergistia | 3 | 0,50 |
Synechococcophycideae | 3 | 2.82 |
Synechococcophycideae | 3 | 2.48 |
|
Thermotogae | 3 | 0,50 |
Synergistia | 3 | 2.82 |
Synergistia | 3 | 2.53 |
|
Flavobacteriia | 3 | 0,06 |
Thermotogae | 3 | 2.80 |
Thermotogae | 3 | 2.53 |
Identification and cluster analysis of
bacterial species in the control, LSIL HPV(+) and HSIL HPV(+)
groups
Analysis of the bacteria species in each of the
three classes of bacteria revealed no cluster formation within the
Gammaproteobacteria class. The further analysis subsequently
focussed only on the Actinobacterium and Bacilli
classes of bacterial species. Regardless of the type of diagnosis,
the bacterial species within the analysed classes formed two
clusters. The results are presented in Tables III and IV (data repository).
 | Table III.The bacteria species of the class
Actinobacterium forming clusters in volunteers and patients
with L-SIL or H-SIL histopathological diagnosis. Gr.- cluster
number, Dist.- Euclidean distance. |
Table III.
The bacteria species of the class
Actinobacterium forming clusters in volunteers and patients
with L-SIL or H-SIL histopathological diagnosis. Gr.- cluster
number, Dist.- Euclidean distance.
|
| Control | L-SIL | H-SIL |
|---|
|
|
|
|
|
|---|
| Bacteria
species | Dist. | Gr. | Bacteria
species | Dist. | Gr. | Bacteria
species | Dist. | Gr. |
|---|
| Gardnerella
vaginalis | 0,00 | 1 | Actinomyces
turicensis | 0,00 | 1 | Gardnerella
vaginalis | 0,00 | 1 |
|
Propionibacterium acnes | 0,00 | 1 | Gardnerella
vaginalis | 0,00 | 1 | Corynebacterium
glaucum | 0,08 | 1 |
| Actinomyces
georgiae | 0,06 | 2 | Actinomyces
georgiae | 0,04 | 2 | Corynebacterium
matruchotii | 0,08 | 1 |
| Actinomyces
neuii | 0,06 | 2 | Actinomyces
neuii | 0,20 | 2 |
Propionibacterium acnes | 0,32 | 1 |
| Actinomyces
odontolyticus | 0,06 | 2 | Actinomyces
odontolyticus | 0,05 | 2 |
Propionibacterium humerusii | 0,15 | 1 |
| Actinomyces
turicensis | 0,06 | 2 | Corynebacterium
amycolatum | 0,04 | 2 | Actinomyces
georgiae | 0,02 | 2 |
| Corynebacterium
amycolatum | 0,02 | 2 | Corynebacterium
aurimucosum | 0,08 | 2 | Actinomyces
neuii | 0,02 | 2 |
| Corynebacterium
aurimucosum | 0,02 | 2 | Corynebacterium
coyleae | 0,06 | 2 | Actinomyces
odontolyticus | 0,06 | 2 |
| Corynebacterium
coyleae | 0,09 | 2 | Corynebacterium
diphtheriae | 0,04 | 2 | Actinomyces
turicensis | 0,02 | 2 |
| Corynebacterium
diphtheriae | 0,02 | 2 | Corynebacterium
glaucum | 0,05 | 2 | Corynebacterium
amycolatum | 0,02 | 2 |
| Corynebacterium
glaucum | 0,02 | 2 | Corynebacterium
glucuronolyticum | 0,04 | 2 | Corynebacterium
aurimucosum | 0,02 | 2 |
| Corynebacterium
glucuronolyticum | 0,06 | 2 | Corynebacterium
jeikeium | 0,04 | 2 | Corynebacterium
coyleae | 0,07 | 2 |
| Corynebacterium
jeikeium | 0,06 | 2 | Corynebacterium
kroppenstedtii | 0,04 | 2 | Corynebacterium
diphtheriae | 0,02 | 2 |
| Corynebacterium
kroppenstedtii | 0,09 | 2 | Corynebacterium
kutscheri | 0,05 | 2 | Corynebacterium
glucuronolyticum | 0,02 | 2 |
| Corynebacterium
kutscheri | 0,25 | 2 | Corynebacterium
lipophiloflavum | 0,05 | 2 | Corynebacterium
jeikeium | 0,020883 | 2 |
| Corynebacterium
lipophiloflavum | 0,02 | 2 | Corynebacterium
matruchotii | 0,13 | 2 | Corynebacterium
kroppenstedtii | 0,02 | 2 |
| Corynebacterium
matruchotii | 0,06 | 2 | Corynebacterium
mucifaciens | 0,05 | 2 | Corynebacterium
kutscheri | 0,07 | 2 |
| Corynebacterium
mucifaciens | 0,02 | 2 | Corynebacterium
pseudogenitalium | 0,06 | 2 | Corynebacterium
lipophiloflavum | 0,16 | 2 |
| Corynebacterium
pseudogenitalium | 0,06 | 2 | Corynebacterium
riegelii | 0,05 | 2 | Corynebacterium
mucifaciens | 0,02 | 2 |
| Corynebacterium
riegelii | 0,02 | 2 | Corynebacterium
sundsvallense | 0,05 | 2 | Corynebacterium
pseudogenitalium | 0,02 | 2 |
| Corynebacterium
sundsvallense | 0,06 | 2 | Corynebacterium
tuberculostearicum | 0,12 | 2 | Corynebacterium
riegelii | 0,02 | 2 |
| Corynebacterium
tuberculostearicum | 0,09 | 2 | Corynebacterium
ureicelerivorans | 0,06 | 2 | Corynebacterium
sundsvallense | 0,06 | 2 |
| Corynebacterium
ureicelerivorans | 0,06 | 2 | Corynebacterium
variabile | 0,06 | 2 | Corynebacterium
tuberculostearicum | 0,02 | 2 |
| Corynebacterium
variabile | 0,06 | 2 |
Propionibacterium acnes | 0,62 | 2 | Corynebacterium
ureicelerivorans | 0,02 | 2 |
|
Propionibacterium avidum | 0,06 | 2 |
Propionibacterium avidum | 0,15 | 2 | Corynebacterium
variabile | 0,02 | 2 |
|
Propionibacterium granulosum | 0,02 | 2 |
Propionibacterium granulosum | 0,05 | 2 |
Propionibacterium avidum | 0,02 | 2 |
|
Propionibacterium humerusii | 0,09 | 2 |
Propionibacterium humerusii | 0,08 | 2 |
Propionibacterium granulosum | 0,02 | 2 |
|
Propionibacterium
microaerophilum | 0,02 | 2 |
Propionibacterium
microaerophilum | 0,05 | 2 |
Propionibacterium
microaerophilum | 0,09 | 2 |
| Streptomyces
lazureus | 0,06 | 2 | Streptomyces
lazureus | 0,02 | 2 | Streptomyces
lazureus | 0,02 | 2 |
 | Table IV.The bacteria species of the class
Bacilli forming clusters in volunteers and patients with L-SIL or
H-SIL histopathological diagnosis. Gr.- cluster number, Dist.-
Euclidean distance. |
Table IV.
The bacteria species of the class
Bacilli forming clusters in volunteers and patients with L-SIL or
H-SIL histopathological diagnosis. Gr.- cluster number, Dist.-
Euclidean distance.
| Control | L-SIL | H-SIL |
|---|
|
|
|
|---|
| Bacteria
species | Dist. | Gr. | Bacteria
species | Dist. | Gr. | Bacteria
species | Dist. | Gr. |
|---|
| Lactobacillus
crispatus | 633,17 | 1 | Lactobacillus
iners | 0,00 | 1 | Lactobacillus
iners | 0,00 | 1 |
| Lactobacillus
iners | 633,17 | 1 | Lactobacillus
acidophilus | 0,00 | 1 | Lactobacillus
acidophilus | 0,00 | 1 |
| Lactobacillus
taiwanensis | 0,00 | 1 | Actinobacillus
parahaemolyticus | 8,19 | 2 | Lactobacillus
crispatus | 0,00 | 1 |
| Actinobacillus
parahaemolyticus | 1,43 | 2 | Actinobacillus
porcinus | 8,17 | 2 | Actinobacillus
parahaemolyticus | 3,84 | 2 |
| Actinobacillus
porcinus | 1,35 | 2 | Actinobacillus
rossii | 8,17 | 2 | Actinobacillus
porcinus | 3,88 | 2 |
| Actinobacillus
rossii | 1,43 | 2 | Alkalibacillus
haloalkaliphilus | 8,07 | 2 | Actinobacillus
rossii | 3,88 | 2 |
| Alkalibacillus
haloalkaliphilus | 1,19 | 2 | Alkalibacillus
salilacus | 7,30 | 2 | Alkalibacillus
haloalkaliphilus | 3,86 | 2 |
| Alkalibacillus
salilacus | 0,47 | 2 | Bacillus
alcaliinulinus | 8,06 | 2 | Alkalibacillus
salilacus | 3,01 | 2 |
| Bacillus
alcaliinulinus | 1,11 | 2 | Bacillus
anthracis | 8,19 | 2 | Bacillus
alcaliinulinus | 3,88 | 2 |
| Bacillus
anthracis | 1,43 | 2 | Bacillus
arbutinivorans | 8,20 | 2 | Bacillus
anthracis | 3,88 | 2 |
| Bacillus
arbutinivorans | 1,43 | 2 | Bacillus
aryabhattai | 8,19 | 2 | Bacillus
arbutinivorans | 3,80 | 2 |
| Bacillus
aryabhattai | 1,35 | 2 | Bacillus
axarquiensis | 8,20 | 2 | Bacillus
aryabhattai | 3,78 | 2 |
| Bacillus
axarquiensis | 1,35 | 2 | Bacillus
azotoformans | 8,16 | 2 | Bacillus
axarquiensis | 3,88 | 2 |
| Bacillus
azotoformans | 1,43 | 2 | Bacillus
benzoevorans | 8,14 | 2 | Bacillus
azotoformans | 3,88 | 2 |
| Bacillus
benzoevorans | 1,43 | 2 | Bacillus
cereus | 8,15 | 2 | Bacillus
benzoevorans | 3,88 | 2 |
| Bacillus
cereus | 1,433 | 2 | Bacillus
flexus | 8,18 | 2 | Bacillus
cereus | 3,84 | 2 |
| Bacillus
flexus | 1,43 | 2 | Bacillus
foraminis | 8,20 | 2 | Bacillus
flexus | 3,88 | 2 |
| Bacillus
foraminis | 1,43 | 2 | Bacillus
fordii | 8,19 | 2 | Bacillus
foraminis | 3,86 | 2 |
| Bacillus
fordii | 1,43 | 2 | Bacillus
fortis | 8,19 | 2 | Bacillus
fordii | 3,88 | 2 |
| Bacillus
fortis | 1,43 | 2 | Bacillus
ginsenggisoli | 8,16 | 2 | Bacillus
fortis | 3,88 | 2 |
| Bacillus
ginsenggisoli | 1,35 | 2 | Bacillus
hackensackii | 8,20 | 2 | Bacillus
ginsenggisoli | 3,88 | 2 |
| Bacillus
hackensackii | 1,43 | 2 | Bacillus
herbersteinensis | 8,19 | 2 | Bacillus
hackensackii | 3,86 | 2 |
| Bacillus
herbersteinensis | 1,35 | 2 | Bacillus
horneckiae | 8,07 | 2 | Bacillus
herbersteinensis | 3,88 | 2 |
| Bacillus
horneckiae | 1,43 | 2 | Bacillus
isabeliae | 8,13 | 2 | Bacillus
horneckiae | 3,86 | 2 |
| Bacillus
isabeliae | 1,43 | 2 | Bacillus
koreensis | 8,19 | 2 | Bacillus
isabeliae | 3,88 | 2 |
| Bacillus
koreensis | 1,43 | 2 | Bacillus
litoralis | 8,17 | 2 | Bacillus
koreensis | 3,88 | 2 |
| Bacillus
litoralis | 1,43 | 2 | Bacillus
mucilaginosus | 8,17 | 2 | Bacillus
litoralis | 3,88 | 2 |
| Bacillus
mucilaginosus | 1,27 | 2 | Bacillus
oleronius | 8,17 | 2 | Bacillus
mucilaginosus | 3,78 | 2 |
| Bacillus
oleronius | 1,43 | 2 | Bacillus
olivae | 8,20 | 2 | Bacillus
oleronius | 3,88 | 2 |
| Bacillus
olivae | 1,27 | 2 | Bacillus
oryzae | 8,15 | 2 | Bacillus
olivae | 3,86 | 2 |
| Bacillus
oryzae | 1,43 | 2 | Bacillus
pseudomegaterium | 8,19 | 2 | Bacillus
oryzae | 3,88 | 2 |
| Bacillus
pseudomegaterium | 1,35 | 2 | Bacillus
sonorensis | 8,15 | 2 | Bacillus
pseudomegaterium | 3,86 | 2 |
| Bacillus
sonorensis | 0,79 | 2 | Bacillus
thermoamylovorans | 8,20 | 2 | Bacillus
sonorensis | 3,88 | 2 |
| Bacillus
thermoamylovorans | 1,43 | 2 | Brevibacillus
brevis | 8,20 | 2 | Bacillus
thermoamylovorans | 3,84 | 2 |
| Brevibacillus
brevis | 1,35 | 2 | Brevibacillus
centrosporus | 8,14 | 2 | Brevibacillus
brevis | 3,80 | 2 |
| Brevibacillus
centrosporus | 1,43 | 2 | Brevibacillus
choshinensis | 8,02 | 2 | Brevibacillus
centrosporus | 3,86 | 2 |
| Brevibacillus
choshinensis | 1,43 | 2 | Brevibacillus
formosus | 7,81 | 2 | Brevibacillus
choshinensis | 3,78 | 2 |
| Brevibacillus
formosus | 1,27 | 2 | Brevibacillus
ginsengisoli | 7,78 | 2 | Brevibacillus
formosus | 3,50 | 2 |
| Brevibacillus
ginsengisoli | 0,95 | 2 | Brevibacillus
invocatus | 8,18 | 2 | Brevibacillus
ginsengisoli | 3,67 | 2 |
| Brevibacillus
invocatus | 1,43 | 2 | Brevibacillus
limnophilus | 8,12 | 2 | Brevibacillus
invocatus | 3,86 | 2 |
| Brevibacillus
limnophilus | 1,35 | 2 | Brevibacillus
panacihumi | 8,07 | 2 | Brevibacillus
limnophilus | 3,78 | 2 |
| Brevibacillus
panacihumi | 0,71 | 2 | Brevibacillus
reuszeri | 8,17 | 2 | Brevibacillus
panacihumi | 3,68 | 2 |
| Brevibacillus
reuszeri | 1,43 | 2 | Geobacillus
thermoglucosidans | 8,18 | 2 | Brevibacillus
reuszeri | 3,88 | 2 |
| Geobacillus
thermoglucosidans | 1,35 | 2 | Lactobacillus
acidifarinae | 8,17 | 2 | Geobacillus
thermoglucosidans | 3,56 | 2 |
| Lactobacillus
acidifarinae | 1,43 | 2 | Lactobacillus
amylolyticus | 8,20 | 2 | Lactobacillus
acidifarinae | 3,88 | 2 |
| Lactobacillus
acidophilus | 19,47 | 2 | Lactobacillus
antri | 19,75 | 2 | Lactobacillus
amylolyticus | 3,86 | 2 |
| Lactobacillus
amylolyticus | 1,35 | 2 | Lactobacillus
apis | 7,11 | 2 | Lactobacillus
antri | 3,54 | 2 |
| Lactobacillus
antri | 1,43 | 2 | Lactobacillus
brantae | 8,17 | 2 | Lactobacillus
apis | 2,78 | 2 |
| Lactobacillus
apis | 118,51 | 2 | Lactobacillus
camelliae | 8,12 | 2 | Lactobacillus
brantae | 3,69 | 2 |
| Lactobacillus
brantae | 0,71 | 2 | Lactobacillus
casei | 8,18 | 2 | Lactobacillus
camelliae | 3,74 | 2 |
| Lactobacillus
camelliae | 0,47 | 2 | Lactobacillus
coleohominis | 8,20 | 2 | Lactobacillus
casei | 3,88 | 2 |
| Lactobacillus
casei | 1,43 | 2 | Lactobacillus
crispatus | 721,14 | 2 | Lactobacillus
coleohominis | 1,50 | 2 |
| Lactobacillus
coleohominis | 1,43 | 2 | Lactobacillus
diolivorans | 8,20 | 2 | Lactobacillus
diolivorans | 3,86 | 2 |
| Lactobacillus
diolivorans | 1,43 | 2 | Lactobacillus
equi | 8,18 | 2 | Lactobacillus
equi | 3,71 | 2 |
| Lactobacillus
equi | 1,43 | 2 | Lactobacillus
equicursoris | 8,13 | 2 | Lactobacillus
equicursoris | 3,84 | 2 |
| Lactobacillus
equicursoris | 1,43 | 2 | Lactobacillus
fabifermentans | 7,99 | 2 | Lactobacillus
fabifermentans | 3,88 | 2 |
| Lactobacillus
fabifermentans | 1,43 | 2 | Lactobacillus
faeni | 7,06 | 2 | Lactobacillus
faeni | 3,27 | 2 |
| Lactobacillus
faeni | 0,23 | 2 | Lactobacillus
farraginis | 8,20 | 2 | Lactobacillus
farraginis | 3,86 | 2 |
| Lactobacillus
farraginis | 1,43 | 2 | Lactobacillus
fermentum | 8,17 | 2 | Lactobacillus
fermentum | 3,49 | 2 |
| Lactobacillus
fermentum | 1,35 | 2 | Lactobacillus
frumenti | 5,83 | 2 | Lactobacillus
frumenti | 3,80 | 2 |
| Lactobacillus
frumenti | 1,43 | 2 | Lactobacillus
gallinarum | 22,36 | 2 | Lactobacillus
gallinarum | 16,20 | 2 |
| Lactobacillus
gallinarum | 2,23 | 2 | Lactobacillus
gasseri | 5,54 | 2 | Lactobacillus
gasseri | 3,57 | 2 |
| Lactobacillus
gasseri | 1,43 | 2 | Lactobacillus
gastricus | 8,20 | 2 | Lactobacillus
gastricus | 3,88 | 2 |
| Lactobacillus
gastricus | 1,35 | 2 | Lactobacillus
gigeriorum | 6,92 | 2 | Lactobacillus
gigeriorum | 2,30 | 2 |
| Lactobacillus
gigeriorum | 0,55 | 2 | Lactobacillus
guizhouensis | 8,20 | 2 | Lactobacillus
guizhouensis | 3,88 | 2 |
| Lactobacillus
guizhouensis | 1,27 | 2 | Lactobacillus
hamsteri | 7,78 | 2 | Lactobacillus
hamsteri | 3,72 | 2 |
| Lactobacillus
hamsteri | 0,07 | 2 | Lactobacillus
helveticus | 12,5 | 2 | Lactobacillus
helveticus | 8,65 | 2 |
| Lactobacillus
helveticus | 2,38 | 2 | Lactobacillus
hilgardii | 8,20 | 2 | Lactobacillus
hilgardii | 3,70 | 2 |
| Lactobacillus
hilgardii | 1,43 | 2 | Lactobacillus
ingluviei | 8,06 | 2 | Lactobacillus
ingluviei | 3,88 | 2 |
| Lactobacillus
ingluviei | 1,43 | 2 | Lactobacillus
intermedius | 22,70 | 2 | Lactobacillus
intermedius | 1,78 | 2 |
| Lactobacillus
intermedius | 7,40 | 2 | Lactobacillus
intestinalis | 8,02 | 2 | Lactobacillus
intestinalis | 3,73 | 2 |
| Lactobacillus
intestinalis | 1,27 | 2 | Lactobacillus
japonicus | 6,42 | 2 | Lactobacillus
japonicus | 3,82 | 2 |
| Lactobacillus
japonicus | 1,59 | 2 | Lactobacillus
jensenii | 268,38 | 2 | Lactobacillus
jensenii | 39,95 | 2 |
| Lactobacillus
jensenii | 1,43 | 2 | Lactobacillus
johnsonii | 113,22 | 2 | Lactobacillus
johnsonii | 10,03 | 2 |
| Lactobacillus
johnsonii | 0,87 | 2 | Lactobacillus
kalixensis | 8,16 | 2 | Lactobacillus
kalixensis | 3,88 | 2 |
| Lactobacillus
kalixensis | 1,43 | 2 | Lactobacillus
kisonensis | 8,20 | 2 | Lactobacillus
kisonensis | 3,86 | 2 |
| Lactobacillus
kisonensis | 1,43 | 2 | Lactobacillus
kitasatonis | 5,94 | 2 | Lactobacillus
kitasatonis | 0,56 | 2 |
| Lactobacillus
kitasatonis | 0,00 | 2 | Lactobacillus
letivazi | 7,74 | 2 | Lactobacillus
letivazi | 3,65 | 2 |
| Lactobacillus
letivazi | 1,11 | 2 | Lactobacillus
manihotivorans | 8,20 | 2 | Lactobacillus
manihotivorans | 3,84 | 2 |
| Lactobacillus
manihotivorans | 1,43 | 2 | Lactobacillus
mucosae | 8,19 | 2 | Lactobacillus
mucosae | 3,81 | 2 |
| Lactobacillus
mucosae | 1,43 | 2 | Lactobacillus
oris | 6,08 | 2 | Lactobacillus
oris | 3,73 | 2 |
| Lactobacillus
oris | 1,43 | 2 | Lactobacillus
paracasei | 8,19 | 2 | Lactobacillus
paracasei | 3,88 | 2 |
| Lactobacillus
paracasei | 1,35 | 2 | Lactobacillus
parafarraginis | 8,20 | 2 | Lactobacillus
parafarraginis | 3,86 | 2 |
| Lactobacillus
parafarraginis | 1,43 | 2 | Lactobacillus
parakefiri | 8,19 | 2 | Lactobacillus
parakefiri | 3,76 | 2 |
| Lactobacillus
parakefiri | 1,43 | 2 | Lactobacillus
paraplantarum | 7,97 | 2 | Lactobacillus
paraplantarum | 3,88 | 2 |
| Lactobacillus
paraplantarum | 1,43 | 2 | Lactobacillus
pentosus | 8,89 | 2 | Lactobacillus
pentosus | 3,66 | 2 |
| Lactobacillus
pentosus | 1,03 | 2 | Lactobacillus
plantarum | 7,78 | 2 | Lactobacillus
plantarum | 3,69 | 2 |
| Lactobacillus
plantarum | 1,03 | 2 | Lactobacillus
reuteri | 7,26 | 2 | Lactobacillus
reuteri | 3,88 | 2 |
| Lactobacillus
reuteri | 1,43 | 2 | Lactobacillus
rhamnosus | 8,04 | 2 | Lactobacillus
rhamnosus | 3,63 | 2 |
| Lactobacillus
rhamnosus | 0,00 | 2 | Lactobacillus
ruminis | 8,20 | 2 | Lactobacillus
ruminis | 3,86 | 2 |
| Lactobacillus
ruminis | 1,43 | 2 | Lactobacillus
salivarius | 8,20 | 2 | Lactobacillus
salivarius | 3,77 | 2 |
| Lactobacillus
salivarius | 1,43 | 2 | Lactobacillus
senmaizukei | 7,43 | 2 | Lactobacillus
senmaizukei | 3,27 | 2 |
| Lactobacillus
senmaizukei | 0,79 | 2 | Lactobacillus
siliginis | 8,11 | 2 | Lactobacillus
siliginis | 3,86 | 2 |
| Lactobacillus
siliginis | 1,35 | 2 | Lactobacillus
similis | 8,20 | 2 | Lactobacillus
similis | 3,76 | 2 |
| Lactobacillus
similis | 1,43 | 2 | Lactobacillus
suebicus | 8,18 | 2 | Lactobacillus
suebicus | 3,84 | 2 |
| Lactobacillus
suebicus | 1,43 | 2 | Lactobacillus
taiwanensis | 477,97 | 2 | Lactobacillus
taiwanensis | 156,77 | 2 |
| Lactobacillus
thailandensis | 1,35 | 2 | Lactobacillus
thailandensis | 8,15 | 2 | Lactobacillus
thailandensis | 3,80 | 2 |
| Lactobacillus
tucceti | 0,87 | 2 | Lactobacillus
tucceti | 7,80 | 2 | Lactobacillus
tucceti | 3,30 | 2 |
| Lactobacillus
ultunensis | 48,75 | 2 | Lactobacillus
ultunensis | 48,09 | 2 | Lactobacillus
ultunensis | 45,05 | 2 |
| Lactobacillus
vaginalis | 1,43 | 2 | Lactobacillus
vaginalis | 6,75 | 2 | Lactobacillus
vaginalis | 3,73 | 2 |
| Lactobacillus
versmoldensis | 1,35 | 2 | Lactobacillus
versmoldensis | 8,18 | 2 | Lactobacillus
versmoldensis | 3,88 | 2 |
| Lactobacillus
zeae | 0,79 | 2 | Lactobacillus
zeae | 8,20 | 2 | Lactobacillus
zeae | 3,86 | 2 |
| Lentibacillus
kapialis | 0,47 | 2 | Lentibacillus
kapialis | 7,82 | 2 | Lentibacillus
kapialis | 3,48 | 2 |
| Lentibacillus
salinarum | 1,43 | 2 | Lentibacillus
salinarum | 8,15 | 2 | Lentibacillus
salinarum | 3,81 | 2 |
| Lysinibacillus
boronitolerans | 1,43 | 2 | Lysinibacillus
boronitolerans | 8,17 | 2 | Lysinibacillus
boronitolerans | 3,88 | 2 |
| Lysinibacillus
cresolivorans | 0,87 | 2 | Lysinibacillus
cresolivorans | 8,12 | 2 | Lysinibacillus
cresolivorans | 3,66 | 2 |
| Lysinibacillus
fusiformis | 1,35 | 2 | Lysinibacillus
fusiformis | 8,20 | 2 | Lysinibacillus
fusiformis | 3,88 | 2 |
| Lysinibacillus
parviboronicapiens | 1,43 | 2 | Lysinibacillus
parviboronicapiens | 8,12 | 2 | Lysinibacillus
parviboronicapiens | 3,78 | 2 |
| Lysinibacillus
xylanilyticus | 1,19 | 2 | Lysinibacillus
xylanilyticus | 8,19 | 2 | Lysinibacillus
xylanilyticus | 3,68 | 2 |
| Paenibacillus
caespitis | 1,43 | 2 | Paenibacillus
caespitis | 8,20 | 2 | Paenibacillus
caespitis | 3,84 | 2 |
| Paenibacillus
castaneae | 1,43 | 2 | Paenibacillus
castaneae | 8,21 | 2 | Paenibacillus
castaneae | 3,84 | 2 |
| Paenibacillus
cellulosilyticus | 1,43 | 2 | Paenibacillus
cellulosilyticus | 8,17 | 2 | Paenibacillus
cellulosilyticus | 3,88 | 2 |
| Paenibacillus
cellulositrophicus | 1,35 | 2 | Paenibacillus
cellulositrophicus | 8,185 | 2 | Paenibacillus
cellulositrophicus | 3,86 | 2 |
| Paenibacillus
cookii | 1,43 | 2 | Paenibacillus
cookii | 8,17 | 2 | Paenibacillus
cookii | 3,88 | 2 |
| Paenibacillus
ehimensis | 1,35 | 2 | Paenibacillus
ehimensis | 8,16 | 2 | Paenibacillus
ehimensis | 3,86 | 2 |
| Paenibacillus
elgii | 1,43 | 2 | Paenibacillus
elgii | 8,20 | 2 | Paenibacillus
elgii | 3,86 | 2 |
| Paenibacillus
filicis | 1,35 | 2 | Paenibacillus
filicis | 8,21 | 2 | Paenibacillus
filicis | 3,88 | 2 |
| Paenibacillus
forsythiae | 1,43 | 2 | Paenibacillus
forsythiae | 8,21 | 2 | Paenibacillus
forsythiae | 3,86 | 2 |
| Paenibacillus
gansuensis | 1,43 | 2 | Paenibacillus
gansuensis | 8,21 | 2 | Paenibacillus
gansuensis | 3,86 | 2 |
| Paenibacillus
ginsengagri | 1,43 | 2 | Paenibacillus
ginsengagri | 8,17 | 2 | Paenibacillus
ginsengagri | 3,81 | 2 |
| Paenibacillus
jamilae | 1,43 | 2 | Paenibacillus
jamilae | 8,21 | 2 | Paenibacillus
jamilae | 3,80 | 2 |
| Paenibacillus
lactis | 1,43 | 2 | Paenibacillus
lactis | 8,21 | 2 | Paenibacillus
lactis | 3,86 | 2 |
| Paenibacillus
macerans | 1,43 | 2 | Paenibacillus
macerans | 8,17 | 2 | Paenibacillus
macerans | 3,88 | 2 |
| Paenibacillus
mendelii | 1,43 | 2 | Paenibacillus
mendelii | 8,21 | 2 | Paenibacillus
mendelii | 3,86 | 2 |
| Paenibacillus
motobuensis | 1,43 | 2 | Paenibacillus
motobuensis | 8,14 | 2 | Paenibacillus
motobuensis | 3,88 | 2 |
| Paenibacillus
naphthalenovorans | 1,43 | 2 | Paenibacillus
naphthalenovorans | 8,18 | 2 | Paenibacillus
naphthalenovorans | 3,88 | 2 |
| Paenibacillus
ourofinensis | 1,03 | 2 | Paenibacillus
ourofinensis | 8,07 | 2 | Paenibacillus
ourofinensis | 3,84 | 2 |
| Paenibacillus
panacisoli | 1,43 | 2 | Paenibacillus
panacisoli | 8,17 | 2 | Paenibacillus
panacisoli | 3,81 | 2 |
| Paenibacillus
pini | 1,35 | 2 | Paenibacillus
pini | 8,20 | 2 | Paenibacillus
pini | 3,88 | 2 |
| Paenibacillus
pocheonensis | 1,43 | 2 | Paenibacillus
pocheonensis | 8,20 | 2 | Paenibacillus
pocheonensis | 3,86 | 2 |
| Paenibacillus
polymyxa | 1,43 | 2 | Paenibacillus
polymyxa | 8,20 | 2 | Paenibacillus
polymyxa | 3,80 | 2 |
| Paenibacillus
residui | 1,43 | 2 | Paenibacillus
residui | 8,18 | 2 | Paenibacillus
residui | 3,88 | 2 |
| Paenibacillus
vortex | 1,43 | 2 | Paenibacillus
vortex | 8,17 | 2 | Paenibacillus
vortex | 3,88 | 2 |
| Paenibacillus
woosongensis | 1,11 | 2 | Paenibacillus
woosongensis | 8,07 | 2 | Paenibacillus
woosongensis | 3,71 | 2 |
| Paenibacillus
xinjiangensis | 1,43 | 2 | Paenibacillus
xinjiangensis | 8,19 | 2 | Paenibacillus
xinjiangensis | 3,80 | 2 |
| Pontibacillus
halophilus | 1,27 | 2 | Pontibacillus
halophilus | 8,02 | 2 | Pontibacillus
halophilus | 3,68 | 2 |
| Pontibacillus
marinus | 1,35 | 2 | Pontibacillus
marinus | 8,19 | 2 | Pontibacillus
marinus | 3,88 | 2 |
| Streptococcus
agalactiae | 1,11 | 2 | Streptococcus
agalactiae | 97,05 | 2 | Streptococcus
agalactiae | 3,65 | 2 |
| Streptococcus
alactolyticus | 1,43 | 2 | Streptococcus
alactolyticus | 8,18 | 2 | Streptococcus
alactolyticus | 3,88 | 2 |
| Streptococcus
anginosus | 1,27 | 2 | Streptococcus
anginosus | 10,85 | 2 | Streptococcus
anginosus | 25,55 | 2 |
| Streptococcus
australis | 1,43 | 2 | Streptococcus
australis | 8,16 | 2 | Streptococcus
australis | 3,88 | 2 |
| Streptococcus
bovis | 1,27 | 2 | Streptococcus
bovis | 7,46 | 2 | Streptococcus
bovis | 3,84 | 2 |
| Streptococcus
cristatus | 1,43 | 2 | Streptococcus
cristatus | 8,17 | 2 | Streptococcus
cristatus | 3,8186 | 2 |
| Streptococcus
fryi | 1,35 | 2 | Streptococcus
fryi | 8,07 | 2 | Streptococcus
fryi | 3,80 | 2 |
| Streptococcus
gallinaceus | 1,43 | 2 | Streptococcus
gallinaceus | 8,20 | 2 | Streptococcus
gallinaceus | 3,80 | 2 |
| Streptococcus
gordonii | 1,43 | 2 | Streptococcus
gordonii | 8,19 | 2 | Streptococcus
gordonii | 3,88 | 2 |
| Streptococcus
infantis | 1,43 | 2 | Streptococcus
infantis | 7,96 | 2 | Streptococcus
infantis | 3,80 | 2 |
| Streptococcus
intermedius | 1,43 | 2 | Streptococcus
intermedius | 8,15 | 2 | Streptococcus
intermedius | 3,88 | 2 |
| Streptococcus
macedonicus | 1,43 | 2 | Streptococcus
macedonicus | 7,97 | 2 | Streptococcus
macedonicus | 3,88 | 2 |
| Streptococcus
milleri | 1,35 | 2 | Streptococcus
milleri | 8,08 | 2 | Streptococcus
milleri | 19,66 | 2 |
| Streptococcus
mutans | 1,43 | 2 | Streptococcus
mutans | 7,98 | 2 | Streptococcus
mutans | 3,88 | 2 |
| Streptococcus
oligofermentans | 1,43 | 2 | Streptococcus
oligofermentans | 8,13 | 2 | Streptococcus
oligofermentans | 3,88 | 2 |
| Streptococcus
oralis | 1,43 | 2 | Streptococcus
oralis | 8,09 | 2 | Streptococcus
oralis | 3,80 | 2 |
| Streptococcus
orisratti | 1,43 | 2 | Streptococcus
orisratti | 10,78 | 2 | Streptococcus
orisratti | 3,84 | 2 |
| Streptococcus
parasanguinis | 1,43 | 2 | Streptococcus
parasanguinis | 8,18 | 2 | Streptococcus
parasanguinis | 3,88 | 2 |
| Streptococcus
pasteuri | 1,43 | 2 | Streptococcus
pasteuri | 8,18 | 2 | Streptococcus
pasteuri | 3,88 | 2 |
| Streptococcus
pseudopneumoniae | 1,27 | 2 | Streptococcus
pseudopneumoniae | 7,73 | 2 | Streptococcus
pseudopneumoniae | 3,88 | 2 |
| Streptococcus
sanguinis | 1,43 | 2 | Streptococcus
sanguinis | 8,19 | 2 | Streptococcus
sanguinis | 3,86 | 2 |
| Streptococcus
thermophilus | 1,35 | 2 | Streptococcus
thermophilus | 8,20 | 2 | Streptococcus
thermophilus | 3,88 | 2 |
| Streptococcus
tigurinus | 1,35 | 2 | Streptococcus
tigurinus | 7,82 | 2 | Streptococcus
tigurinus | 3,78 | 2 |
| Streptococcus
vestibularis | 1,27 | 2 | Streptococcus
vestibularis | 8,03 | 2 | Streptococcus
vestibularis | 3,47 | 2 |
| Ureibacillus
thermophilus | 1,43 | 2 | Ureibacillus
thermophilus | 8,19 | 2 | Ureibacillus
thermophilus | 3,86 | 2 |
| Virgibacillus
byunsanensis | 1,43 | 2 | Virgibacillus
byunsanensis | 8,20 | 2 | Virgibacillus
byunsanensis | 3,86 | 2 |
| Virgibacillus
salexigens | 1,43 | 2 | Virgibacillus
salexigens | 8,20 | 2 | Virgibacillus
salexigens | 3,84 | 2 |
| Viridibacillus
arvi | 0,87 | 2 | Viridibacillus
arvi | 8,18 | 2 | Viridibacillus
arvi | 3,67 | 2 |
| Viridibacillus
neidei | 1,35 | 2 | Viridibacillus
neidei | 8,18 | 2 | Viridibacillus
neidei | 3,88 | 2 |
The Actinobacterium class included
Gardnerella vaginalis and Propionibacterium acnes
identified in the CST healthy women; Gardnerella vaginalis
and Actinomyces turicensis in the CST women diagnosed with
LSIL, and Gardnerella vaginalis, Corynebacterium glaucum,
Corynebacterium matruchotii, Propionibacterium acnes and
Propionibacterium humerusii in women diagnosed with HSIL,
which formed clusters separate from the other bacterial species
(Table III, data repository).
The cluster analysis of the Bacilli bacterial
species revealed the presence of a CST subpopulation, which in the
healthy women consisted of Lactobacillus crispatus,
Lactobacillus iners and Lactobacillus taiwanensis. In
women diagnosed with LSIL it included Lactobacillus iners
and Lactobacillus acidophilus, and in women diagnosed with
HSIL it included Lactobacillus iners, Lactobacillus
acidophilus and Lactobacillus crispatus (Table IV, data repository). For further
analysis, the levels of bacterial characteristics were determined
in each of the analysed patient groups (Table V).
 | Table V.Analysis of the selected bacteria
species frequency in control, LSIL and HSIL diagnosed patients.
Frequencies >10 have been highlighted. |
Table V.
Analysis of the selected bacteria
species frequency in control, LSIL and HSIL diagnosed patients.
Frequencies >10 have been highlighted.
| Species | Control | LSIL | HSIL |
|---|
| Actinomyces
turicensis | 0 | 0.63 | 0 |
| Corynebacterium
glaucum | 0.08 | 0 | 0.19 |
| Corynebacterium
matruchotti | 0 | 0.07 | 0.19 |
| Gardnerella
vaginalis | 11.47 | 46.93 | 304.49 |
| Lactobacillus
acidophilus | 20.87 | 1273.62 | 1193.89 |
| Lactobacillus
crispatus | 2506.01 | 405.03 | 245.87 |
| Lactobacillus
iners | 3772.35 | 3183.67 | 1217.21 |
| Lactobacillus
taiwanensis | 320.69 | 410.57 | 116.49 |
|
Propionibacterium acnes | 0.88 | 0.42 | 0.48 |
|
Propionibacterium humerusii | 0.16 | 0.13 | 0.14 |
The analysis of the bacterial classes indicated that
CST cervical swabs of the female patients were colonised by
Lactobacillus crispatus, Lactobacillus iners and
Lactobacillus taiwanensis, however, Gardnerella
vaginalis and Lactobacillus acidophilus were almost not
identified. In the CST patients diagnosed with LSIL, the
predominant types of bacteria were Lactobacillus acidophilus
and Lactobacillus iners, while Lactobacillus
crispatus frequency was lower than in the control group. In CST
patients diagnosed with HSIL, high abundance of Gardnerella
vaginalis, Lactobacillus acidophilus was identified, however,
Lactobacillus taiwanensis, Lactobacillus iners and
Lactobacillus crispatus frequencies were lower than in the
control group. The level of Lactobacillus acidophilus in the
CST patients diagnosed with LSIL swabs were compared with swabs
taken from women with HSIL.
Discussion
It has been indicated that the introduction of
Next-Generation Sequencing (NGS) into research allowed for the
identification of specific ecological niches for microorganisms
within living organisms. This was possible due to the amplification
and parallel sequencing of gene fragments, which were highly
conserved among microorganisms, the most common being the 16S rRNA
subunit, as well as RAD51 recombinase and inhibin subunit a
(12). Highly conserved regions of
the 16S rRNA gene (V1-V9) have been reported to allow for
phylogenetic and taxonomic characterisation of the analysed
microbial communities (12). In the
present study, the V4 hypervariable regions of 16S rRNA were used,
which allowed for the identification of 3 CSTs of HSIL HPV(+), LSIL
HPV(+) and the healthy group HPV(−).
In addition, >70 classes of bacteria were
identified and the analysis of their frequencies demonstrated that
cervical swabs of flora obtained from the volunteers and women
diagnosed with LSIL HPV(+) were predominantly composed of the
Bacilli class. The presence of Gammaproteobacteria
and Actinobacterium classes in patients with HSIL HPV(+)
were also detected. Further analysis revealed no cluster formation
within the Gammaproteobacteria class and, therefore, this
class was excluded from the present study.
Further analysis focussed only on the
Actinobacterium and Bacilli classes of bacterial
species. Regardless of the type of diagnosis, the bacterial species
within the analysed classes only formed two clusters. It was
observed that swabs from healthy women were characterised by an
increased level of Lactobacillus crispatus, Lactobacillus
iners and Lactobacillus taiwanensis. However,
Gardnerella vaginalis and Lactobacillus acidophilus
were absent. In the CST patients diagnosed with LSIL HPV(+) the
predominant types of bacteria were Lactobacillus acidophilus
and Lactobacillus iners, and there was no presence of
Lactobacillus crispatus detected. The CST cervical swabs
obtained from women with HSIL HPV(+) were rich in Gardnerella
vaginalis and Lactobacillus acidophilus, while
Lactobacillus taiwanensis, Lactobacillus iners and
Lactobacillus crispatus were not detected. However, similar
levels of Lactobacillus acidophilus were identified in the
control and LSIL HPV(+) groups.
In the majority of healthy women in the present
study, Lactobacillus crispatus, Lactobacillus iners and
Lactobacillus taiwanensis were the dominant cervical
microbiota. According to previous studies, these species were also
reported to be dominant in the vaginal microbiota of Asian women
(6,13). The women with the highest risk of HSIL
HPV(+) indicated low levels of Lactobacillus crispatus,
Lactobacillus iners and Lactobacillus taiwanensis. It
has been reported that hydrogen peroxide-producing Lactobacilli are
present in 96% of women with a normal vaginal bacterial community
(4). In addition, species in the
Lactobacillus genus have been reported to maintain a low pH
by producing lactic acid (4). It has
been reported that Lactobacillus crispatus was associated
with a low vaginal pH compared with Lactobacillus iners,
suggesting that these two species differ in ecological function
(14). The cervical swabs obtained
from healthy women consisted mainly of Lactobacillus
bacteria. Due to their presence, the physiological pH value
(3.6–4.5) was achieved by fermentation of epithelial glycogen. In
addition, it has been reported that Lactobacillus bacteria
have a high adherence potential, which allows them to adhere
tightly to the vaginal epithelium and cover its surface, protecting
the vagina from colonisation by pathogenic microorganisms (15–17). A
number of Lactobacillus species can form biofilms and
produce bacteriocins and hydrogen peroxide, which together have
been reported to inhibit the development of undesirable anaerobes
in the vagina (15,16). Lactobacillus iners has been
previously reported to become a predominant part of the microbial
community in the vaginal microbiota transition between abnormal and
normal states (17). HPV infection
can alter the mucosal metabolism and host immunity, and can induce
changes in the vaginal microbiota (18).
It has been reported that microbe-induced
inflammation can contribute to cervical cancer by stimulating the
production of specific cytokines and chemokines, which promote
proliferation and/or inhibit apoptosis (18,19).
Microbiota have been demonstrated to increase, as well as decrease
susceptibility to HPV infection (18,19). An
association between vaginal microbiota and HPV infection was
previously described by Gao et al (19). They observed a significant presence of
Lactobacillus species, including Lactobacillus
gallinarum, Lactobacillus iners and Lactobacillus
gasseri in all women as well as Lactobacillus gasseri
and Gardnerella vaginalis in HPV(+) women. The reduced
population of the Lactobacillus species and the presence of
Fusobacteria species in HPV(+) patients was also observed by
Lee et al (6). The present
study demonstrated that bacterial communities in the cervix are
more complex than previously thought. The analyses suggest an
association between HPV infection and decreased abundance of
Lactobacillus species and increased abundance of
Gammaproteobacteria anaerobes. The aforementioned results
are similar to those reported by Lee et al (6) and Dareng et al (8). The changes to the ‘core microbiome’ may
be associated with changes in human health and risk of exposure to
HPV infection and cervical cancer development.
In summary, analysis of the association between the
occurrence of bacterial species and the histopathological diagnosis
in the analysed population revealed that the vaginal microbiota may
be clustered into two groups. Cluster 1 was predominantly affected
by Lactobacillus crispatus, Lactobacillus iners and
Lactobacillus taiwanensis and Cluster 2 predominated by
Gardnerella vaginalis. The frequency of Lactobacillus
acidophilus was identical in the clustered groups. The proposed
mechanism of a cervical cancer formation may start with a sexually
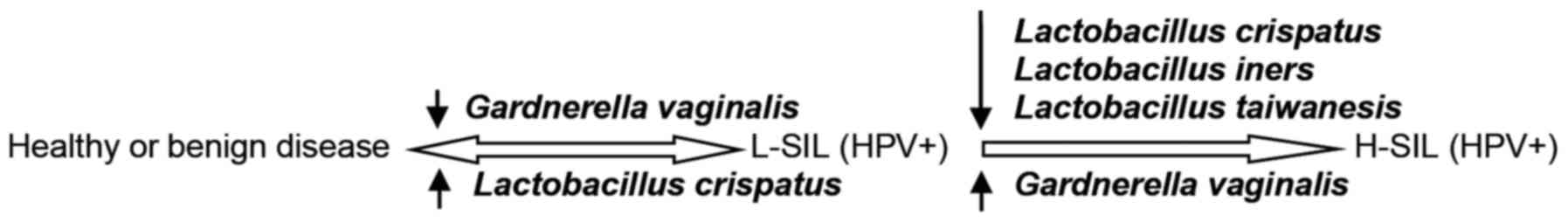
transmitted carcinogenic HPV infection (Fig. 1).
The fastest rate of HPV clearance may occur in women
with Cluster 1 bacteria as the dominant microbiota as >50% of
all infections were cleared within a year. The lowest clearance
rate may occur in women classified with Cluster 2 dominant
microbiota. In the aforementioned patients the chance of HSIL(+)
development gradually increases, representing the growth of a
clonal high-grade lesion up to a number of years following HPV
infection. It was recently demonstrated that Gardnerella
vaginalis, a dominant Cluster 2 bacterium, was able to adhere
to and displace precoated protective Lactobacilli from
vaginal epithelial cells, while other BV-associated anaerobes,
including Atopobium vaginae, were less virulent (20).
The findings of the present study demonstrated a
possible interaction between bacterial flora and HPV infection, as
well as an association between this interaction and clinical
cervical neoplasia. It was observed that bacterial dysbiosis,
characterized by a predominance of Gardnerella vaginalis and
a concomitant paucity of Lactobacillus crispatus, Lactobacillus
iners and Lactobacillus taiwanensis, may be an
HPV-dependent cofactor for cervical neoplasia development. However,
without continuous observation it is difficult to confirm that
microbiota dysbiosis contributes to HPV infection and
carcinogenesis. Future researches are required to confirm the
results of 16S rRNA sequencing and determine that microflora
dysbiosis may be associated with HPV-induced cervical
carcinogenicity.
Acknowledgements
Not applicable.
Funding
The present study was funded by grants from the
Medical University of Lublin (Lublin, Poland; grant nos., DS 120,
DS 128 and MB 128).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request. The datasets presented in Tables III and IV can be found online at the following
webpages: http://www.katbiolkom.ump.edu.pl/wp-content/uploads/2018/07/table_III.docx
and http://www.katbiolkom.ump.edu.pl/wp-content/uploads/2018/07/table_IV.doc
× (The repository of the University of Medical Sciences, Poznan,
Poland).
Authors' contributions
WK conceived the study, collected samples, wrote the
materials and methods and edited the manuscript. MWC analyzed the
data and wrote the results and discussion sections. JK conceived
the study, collected samples and contributed resources. DK
performed laboratory assays. WW conceived the study, prepared
figures and wrote background information. AK conceived the study,
collected samples, contributed resources and approved the final
draft. AGJ conceived the study, contributed resources, collected
samples and approved the final draft.
Ethics approval and consent to
participate
The investigations were approved by the Ethics
Committee of the Medical University of Lublin (Lublin, Poland;
Resolution of the Bioethics Committee; approval no: 0254/30/2002.
Written informed consent was obtained from all individuals, and the
research was performed by the principles of the Helsinki
Declaration.
Patient consent for publication
Study participants provided written informed consent
for the publication of any data and associated images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wheeler CM: The natural history of
cervical human papillomavirus infections and cervical cancer: Gaps
in knowledge and future horizons. Obstet Gynecol Clin North Am.
40:165–76. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Oh HY, Kim BS, Seo SS, Kong JS, Lee JK,
Park SY, Hong KM, Kim HK and Kim MK: The association of uterine
cervical microbiota with an increased risk for cervical
intraepithelial neoplasia in Korea. Clin Microbiol Infect.
21:674.e1–e9. 2015. View Article : Google Scholar
|
|
3
|
Polatti F: Bacterial vaginosis, Atopobium
vaginae and nifuratel. Curr Clin Pharmacol. 7:36–40. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nam H, Whang K and Lee Y: Analysis of
vaginal lactic acid producing bacteria in healthy women. J
Microbiol. 45:515–520. 2007.PubMed/NCBI
|
|
5
|
Hillier SL, Critchlow CW, Stevens CE,
Roberts MC, Wolner-Hanssen P, Eschenbach DA and Holmes KK:
Microbiological, epidemiological and clinical correlates of vaginal
colonisation by Mobiluncus species. Genitourin Med. 67:26–31.
1991.PubMed/NCBI
|
|
6
|
Lee JE, Lee S, Lee H, Song YM, Lee K, Han
MJ, Sung J and Ko G: Association of the vaginal microbiota with
human papillomavirus infection in a Korean twin cohort. PLoS One.
8:e635142013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gillet E, Meys JF, Verstraelen H, Bosire
C, De Sutter P, Temmerman M and Broeck DV: Bacterial vaginosis is
associated with uterine cervical human papillomavirus infection: A
meta-analysis. BMC Infect Dis. 11:102011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dareng EO, Ma B, Famooto AO,
Akarolo-Anthony SN, Offiong RA, Olaniyan O, Dakum PS, Wheeler CM,
Fadrosh D, Yang H, et al: Prevalent high-risk HPV infection and
vaginal microbiota in Nigerian women. Epidemiol Infect.
144:123–137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
The 1988 Bethesda System for reporting
cerval/vaginal cytologic diagnoses: Developed and approved at the
National Cancer Institute workshop in Bethesda, MD, December 12-13,
1988. Diagn Cytopathol. 5:331–334. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Manos MM, Ting Y, Wright DK, Lewis AJ,
Broker TR and Wolinsky SM: The use of polymerase chain reaction
amplification for the detection of genital human papilloma viruses.
Cancer Cell. 7:209–214. 1989.
|
|
11
|
Fadrosh DW, Ma B, Gajer P, Sengamalay N,
Ott S, Brotman RM and Ravel J: An improved dual-indexing approach
for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq
platform. Microbiome. 2:62014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu D, Wu M, Halpern A, Rusch DB, Yooseph
S, Frazier M, Venter JC and Eisen JA: Stalking the fourth domain in
metagenomic data: Searching for, discovering, and interpreting
novel, deep branches in marker gene phylogenetic trees. PLoS One.
6:e180112011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ravel J, Gajer P, Abdo Z, Schneider GM,
Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO,
et al: Vaginal microbiome of reproductive-age women. Proc Natl Acad
Sci USA. 108:4680–4687. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hummelen R, Fernandes AD, Macklaim JM,
Dickson RJ, Changalucha J, Gloor GB and Reid G: Deep sequencing of
the vaginal microbiota of women with HIV. PLoS One. 5:e120782010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Petricevic L, Domig KJ, Nierscher FJ,
Sandhofer MJ, Fidesser M, Krondorfer I, Husslein P, Kneifel W and
Kiss H: Characterisation of the vaginal Lactobacillus microbiota
associated with preterm delivery. Sci Rep. 4:51362014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mitra A, MacIntyre DA, Marchesi JR, Lee
YS, Bennett PR and Kyrgiou M: The vaginal microbiota, human
papillomavirus infection and cervical intraepithelial neoplasia:
What do we know and where are we going next? Microbiome. 4:582016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jakobsson T and Forsum U: Lactobacillus
iners: A marker of changes in the vaginal flora? J Clin Microbiol.
45:31452007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Scott M, Stites DP and Moscicki AB: Th1
cytokine patterns in cervical human papillomavirus infection. Clin
Diagn Lab Immunol. 6:751–755. 1999.PubMed/NCBI
|
|
19
|
Gao W, Weng J, Gao Y and Chen X:
Comparison of the vaginal microbiota diversity of women with and
without human papillomavirus infection: A cross-sectional study.
BMC Infec Dis. 13:2712013. View Article : Google Scholar
|
|
20
|
Menard JP, Fenollar F, Henry M, Bretelle F
and Raoult D: Molecular quantification of Gardnerella vaginalis and
Atopobium vaginae loads to predict bacterial vaginosis. Clin Infect
Dis. 47:33–43. 2008. View
Article : Google Scholar : PubMed/NCBI
|