Introduction
It is generally accepted that chronic myelogenous
leukemia (CML) is caused by a reciprocal translocation of
chromosomes 9 and 22 termed the Philadelphia (Ph) chromosome which
results in the formation of the BCR-ABL fusion gene in primitive
hematopoietic cells (HCs) (1). It has
previously been reported that the bcr-abl fusion gene initially
appears in one single HC, which subsequently induces a growth
advantage for the cell and leads to excess multiplication (2). Over-accumulation of CML-HCs produces
excessive amounts of granulocyte colony-stimulating factor, which
downregulates the expression of C-X-C motif chemokine ligand 12
(CXCL12) in the bone marrow microenvironment (BMM) of CML. This
downregulation of CXCL12 results in a reduction of the homing and
retention of HCs and an increase in the number of CML-HCs in
peripheral blood (3). Due to the
genome instability in CML, CML-HCs are affected by accumulation of
point mutations and chromosomal aberrations, which eventually
facilitate blastic transformation (4,5).
Within a decade, CML has progressed from a fatal
disease to a disorder that can be controlled by lifelong oral
medication (1). Use of BCR-ABL
tyrosine kinase inhibitors has markedly improved the treatment of
CML (6). However, drug resistance has
become increasingly prominent in recent years (7). Previous studies have focused on drug
combination (8) and protective
effects of the BMM (9,10), whereas the current study aims to
provide a new perspective by investigating the changes that occur
in the CML-BMM.
The BMM not only protects HCs from drugs but also
participates in the development of leukemia (9,10). As one
of the main components of BMM, bone mesenchymal stromal cells
(BMSCs) serve an indispensable role in hematopoiesis regulation
(11). Previous studies have
demonstrated that BMSCs derived from patients with myelodysplastic
syndromes (MDS) and acute myeloid leukemia (AML) exhibit biological
function impairment and a higher inclination to senescence compared
with BMSCs derived from healthy donors (HDs) (12–14). In
addition, genetic aberrations were detected in HCs in approximately
20% of patients with AML and 12% of patients with MDS (15). To the best of our knowledge, previous
research concerning CML has mainly focused on malignant HCs, with
less focus on CML-BMSCs. This prompted the current study to analyze
the biological characteristics and genetic alterations of the
CML-BMM in order to explore its role in hematopoietic
dysfunction.
Materials and methods
Patients and samples
A total of 6 healthy donors (HDs) (3 male, 3 female,
28–44 years of age, with a median age of 39 years) without
malignant hematologic disease and 15 patients with CML diagnosed
according to the 2016 World Health Organization criteria (16), were included in the current study. All
patients and HDs were hospitalized at Fujian Medical University
Union Hospital (Fuzhou, China) from August 2016 to March 2017.
Patient clinical characteristics are summarized in Table I. Fresh heparinized bone marrow
samples were obtained from the anterior superior iliac spine based
on donors written informed consent. The current study was approved
by the Ethical Committee of Fujian Medical University Union
Hospital.
 | Table I.Demographics and clinical
characteristics of patients with chronic myelogenous leukemia
included in the current study. |
Table I.
Demographics and clinical
characteristics of patients with chronic myelogenous leukemia
included in the current study.
|
Characteristics | No. | % |
|---|
| Age, years | 45 |
|
|
Range | 24–67 |
|
| Sex |
|
|
|
Male | 8 | 53.3 |
|
Female | 7 | 46.7 |
| Karyotype in
HCs |
|
|
|
Normal | 0 | 0 |
|
Aberrant | 15 | 100.0 |
| FISH signals in
HC |
|
|
|
Normal | 0 | 0 |
|
Aberrant | 15 | 100.0 |
| Median WBC,
×109/l | 172.95 |
|
|
Range | 17.16–653.12 |
|
| Median ANC,
×109/l | 94.17 |
|
|
Range | 14.71–575.98 |
|
| Median Hb,
g/dl | 92.50 |
|
|
Range | 52.00–197.00 |
|
| Median PLTs,
×109/l | 223.50 |
|
|
Range | 14.00–717.00 |
|
Isolation and culture of BMSCs
BMSCs were isolated from bone marrow aspirate by
density gradient centrifugation (500 × g, 30 min at 25°C) using
Ficoll-Paque Plus (TBD, Tianjin, China, http://www.tbdscience.com/). Mononuclear cells were
cultured with Dulbecco's modified Eagle's-low glucose medium (DMEM)
(Hyclone; GE Healthcare, Logan, UT, USA) containing 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) at 37°C under humidified atmosphere of 5% CO2.
After 24–48 h, the culture medium was discarded to remove
non-adherent cells. Half of the medium was renewed every 3–4 days.
When confluency reached 70–80%, the cells were detached with 0.25%
trypsin EDTA (Hyclone; GE Healthcare, Logan, UT, USA) for
subculture.
Cell surface markers of BMSCs
Cell surface antigens were analyzed using a flow
cytometer (Beckman Coulter, Inc., Brea, CA, USA). BMSCs at passage
3 were harvested and adjusted to 1×105 cells per tube. A
total of six tubes were separately resuspended with 100 µl
phosphate buffer solution (PBS) (Hyclone; GE Healthcare) and
labeled with 5 µl monoclonal antibodies per 1×105 cells
for 30 min in the dark at 4°C as follow: anti-CD45-phycoerythrin
(PE) (12-0459-41), anti-CD34-PE (12-0349-41), anti-CD73-PE
(12-0739-41), anti-CD90-PE (12-0909-41), anti-CD105-PE (12-1057-41)
(all monoclonal antibodies 1:20 dilution, eBiosciences; Thermo
Fisher Scientific Inc.). Species and isotype matched antibodies
served as controls. Flow cytometric data analysis was made by
Cytomics FC 500 analysis software (CXP 2.0, Beckman Coulter).
Colony-forming unit fibroblast (CFU-F)
assay
BMSCs at passage 3 were detached with trypsin EDTA
(Hyclone; GE Healthcare) and plated at a low density
~1×102/ml onto a fresh culture dish. The cells were
cultured for 14 days at 37°C under humidified atmosphere of 5%
CO2 in DMEM (Hyclone; GE Healthcare) and half of the
medium was renewed every 4 days. The cells were then fixed with 4%
paraformaldehyde for 20 min and stained with Giemsa for 30 min at
room temperature. The stained cells were observed under a Nikon
7S-100 light microscope (Nikon Corporation, Tokyo, Japan) and
colonies including ≥50 cells were scored as a CFU-F.
Differentiation assay. BMSCs were harvested and
seeded to 6-well culture dishes at a concentration of
2×104/well with DMEM (Hyclone; GE Healthcare). When the
culture reached 80% confluency ~5×105/well, the medium
was replaced with differentiation and induction medium from the
StemPro Adipogenesis Differentiation kit, StemPro Osteogenesis
Differentiation kit and the StemPro Chondrogenesis Differentiation
kit (all kits from Invitrogen; Thermo Fisher Scientific, Inc.) for
2 weeks. The extent of adipogenesis, osteogenesis and
chondrogenesis was observed by staining with oil red O, alizarin
red S and alcian blue, respectively according to the manufacturer's
protocol at room temperature (all dyes from Beijing Solarbio
Science & Technology Co., Ltd., Beijing, China).
Cell Counting Kit-8 (CCK8) assay to
measure cell proliferation
BMSCs in an exponential phase of growth were
collected and seeded into 96-well plates at a concentration of
2×104/well based on the CCK8 assay which was performed
once a day for a total of 6 days. At 4 h after the cells were
adhered, 10 µl CCK8 (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) was added to each well. Following 2 h of
incubation at 37°C under humidified atmosphere of 5%
CO2, densitometry was detected at the wavelength of 450
nm by a microplate reader (BioTek Instruments, Inc., Winooski, VT,
USA).
Senescence associated-β-galactosidase
(SA-β-gal staining assay)
BMSCs were harvested at passage 8 and seeded into a
6-well plate at a concentration of 2×104 per well. An
SA-β-gal staining kit (Beyotime Institute of Biotechnology, Haimen,
China) was used to evaluate the percentage of senescent cells,
according to the manufacturer's protocol. Subsequently, the plates
were incubated for 12 h at 37°C under humidified atmosphere but
without CO2. The percentage of blue-stained senescent
cells per 500 cells was calculated in at least 10 random fields
using a Nikon 7S-100 light microscope.
Long-term culture-initiating cell
(LTC-IC) assay
An LTC-IC assay was conducted to detect the
hematopoietic support ability of BMSCs. Firstly, BMSCs layers were
seeded with 10 µl/ml mitomycin-c (Sigma-Aldrich) for 2 h at 37°C in
humidified atmosphere of 5% CO2. Then, HDs-HCs sorted
using the CD34 MicroBead kit (Miltenyi Biotec Inc., Cambridge, MA,
USA), were seeded onto the BMSC layers with MyeloCult H5100 medium
(Stemcell Technologies, Inc., Vancouver, BC, Canada) for 5 weeks.
Co-cultured CD34+ cells were then collected and cultured
in MethoCult H4431 medium (Stemcell Technologies, Inc.) at a
concentration of 1×103/ml per dish for a further 14
days. Colony forming units of granulocyte/monocyte (CFU-GM) (≥40
cells), colony forming units of erythroid (CFU-E) (≥50 cells) and
burst forming units of erythroid (BFU-E) (≥300 cells) were counted
under the Nikon 7S-100 light microscope.
Conventional cytogenetic (CC) analysis
and fluorescent in situ hybridization (FISH) analysis of HCs and
BMSCs
Cytogenetics of HCs and BMSCs were evaluated by CC
analysis and FISH analysis. Briefly, the cells were maintained at
metaphase by adding colcemide (Gibco; Thermo Fisher Scientific,
Inc.) at a final concentration of 0.2 µg/ml. The air-drying method
was used to prepare the glass slides. CC analysis was carried out
by R banding (17). Karyotypic
abnormalities were described in detail in accordance with the
International System for Human Cytogenetic Nomenclature (2013)
(18). A BCR-ABL probe (GP Medical
Technologies, Ltd., Beijing, China, http://www.gpmedical.com.cn/) was used according to
the manufacturer's protocol to measure BCR-ABL translocations by
FISH analysis. ABL gene on chromosome 22 is marked with the red
signal, BCR gene on chromosome 9 with the green signal, and the
hybridization signal of BCR-ABL fusion gene shows the yellow
signal. The results were detected by an Olympus BX51 fluorescence
microscope (Olympus Corporation, Tokyo, Japan).
Statistical analysis
Quantitative data are presented as mean ± standard
deviation. A two-sided unpaired Student's t-test was applied to
compare two different groups using SPSS 20.0 (IBM Corp., Armonk,
NY, USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
Identification of BMSCs
Samples from HDs and patients with CML were
collected to verify whether isolated cells met the minimal
definition criteria for BMSCs proposed by the International Society
for Cellular Therapy (ISCT) (19). As
demonstrated in Fig. 1A, the isolated
cells from HDs were negative for HC antigens, including CD34 and
CD45 (<10%), but positive for CD73, CD90 and CD105 (>90%),
which was consistent with previous studies (14,20), as
well as CML-BMSCs (data not shown). A CFU-F assay also verified the
isolated cells were BMSCs. As demonstrated in Fig. 1B, one single cell from HDs was able to
form a visible colony at passage 3 and this characteristic could
also be detected at passage 8 (data not shown) as well as CML-BMSCs
(data not shown), indicating the CML-BMSCs did not lose
self-renewal capacity before or at passage 8. A differentiation
assay revealed that HD-BMSCs at passage 3 exhibited the ability to
undergoing multipotential differentiation as it could turned into
three different shapes after being induced by the corresponding
induction differentiation culture conditions as illustrated in
Fig. 1C, which was still maintained
at passage 6 (data not shown). Likewise, multipotential
differentiation ability was also observed in CML-BMSCs (data not
shown).
 | Figure 1.Identification of BMSCs. (A) BMSCs
were negative for CD45 (1.92%) and CD34 (7.21%) but positive for
CD73 (88.54%), CD90 (98.82%) and CD105 (99.58%). (B) By seeding
1,000 cells in a 10 mm dish, BMSCs demonstrated the ability to form
a visible colony unit as indicated by the arrows. (C) BMSCs
exhibited the capacity to differentiate to (C-a) adipogenic cells,
(C-b) osteogenic cells and (C-c) chondrocytes as indicated by the
arrows. The corresponding staining results are presented in in
(Cd-f), (C-d) adipogenic cells stained with oil red O, (C-e)
osteogenic cells stained with alizarin red S and (C-f) chondrocytes
stained with alcian blue are indicated by the arrows. (Cg-i)
Negative controls were undifferentiated BMSCs respectively stained
with (Cg) oil red O, (Ch) alizarin red S and (Ci) alcian blue which
were indicated by the arrows. (D-a) HD-BMSCs and (D-b) CML-BMSCs at
passage 3 exhibited fibroblast-like morphology as indicated by the
arrows. (D-c) Certain HD-BMSCs at passage 5 grew excessively large
and exhibited an irregular shape as indicated by the arrows. (D-d)
Compared with HD-BMSCs, a higher number of CML-BMSCs at passage 5
grew excessively larger and exhibited an irregular shape as
indicated by the arrows. Black lines represent 10 µm.
Magnification, ×100. CON, control; BMSCs, bone mesenchymal stromal
cells; HD, healthy donor; CML, chronic myelogenous leukemia. |
In summary, the adherent cells isolated were BMSCs,
which possessed stable stem cell properties that provided
sufficient reliability for subsequent experimentation.
BMSC culture and morphology
The amplification culture of HD-BMSCs and CML-BMSCs
was measured. It was identified that it took 15 days on average,
with a range of 7–30 days, for primary BMSCs to reach 80%
confluency. Notably, it appeared to be harder for HD-BMSCs to adapt
to culture conditions in vitro compared with CML-BMSCs.
HD-BMSCs required a longer period of time (average of 20 days) to
pass on to the next generation than CML-BMSCs (average of 10 days)
at the first division. Both HD-BMSCs and CML-BMSCs grew at a faster
rate after passage 1 (average of 3–7 days) and then slowed down in
growth at passage 5 (average of 5–14 days).
The current study demonstrated that both HD-BMSCs
and CML-BMSCs exhibited fibroblast-like shape at the early passage
such as passage 3 (Fig. 1D a-b). As
the passages increased, a higher number of CML-BMSCs grew larger
and more irregular in shape compared with HD-BMSCs at passage 5
(Fig. 1D c-d).
Impaired growth kinetics of
CML-BMSCs
To evaluate the growth rate of HD-BMSCs and
CML-BMSCs, a CCK8 assay was performed. Over the 6-day culture
period, CML-BMSCs grew at a significantly slower speed compared
with HD-BMSCs between days 3 and 6 (P<0.05; Fig. 2A). Furthermore, by seeding 200 cells
per dish, a CFU-F assay was performed to assess the self-renewal
ability of BMSCs. CML-BMSCs produced a significantly lower number
of CFU-Fs compared with HD-BMSCs (7.67±1.10 vs. 15.67±0.67;
Fig. 2B).
 | Figure 2.Biological characteristics of BMSCs.
(A) Cell proliferation capacity of CML-BMSCs (n=5) was
significantly impaired compared with that of HD-BMSCs (n=5) on days
3 to 6. (B) Compared with HD-BMSCs (n=6) the CFU-F number of
CML-BMSCs (n=12) was significantly lower (15.67±0.67 vs. 7.67±1.1).
(C) The percentage of senescent CML-BMSCs was significantly higher
compared with that of HD-BMSCs (60.17±2.66 vs. 22.33±1.61%). The
senescent cells were stained blue as indicated by the arrows. Black
lines represent 10 µm. Magnification, ×100. (D) The hematopoietic
support capacity was impaired in CML-BMSCs compared with that of
HD-BMSCs. CFU-GM as indicated by the arrows (left panel) formed by
CD34+ HCs following co-culture with CML-BMSCs was smaller than that
co-culture with HD-BMSCs (the bigger CFU-GM as indicated by the
arrows in right panel). CFU-E as indicated by the arrows appeared
more in CD34+ HCs-CML-BMSCs co-culture system, while BFU-E as
indicated by the arrows appeared more in CD34+ HCs-HD-BMSCs
co-culture system (The picture from left to right top to bottom is
as follows: Smaller CFU-GM, bigger CFU-GM, CFU-E and BFU-E as
indicated by the arrows). The numbers of CFU-GM, CFU-E and BFU-E
formed by CD34+ HCs were decreased when cultured with CML-BMSCs as
opposed to HD-BMSCs (histogram). Black lines represent 10 µm.
Magnification, ×400. Data are presented as the mean ± standard
deviation. *P<0.05, **P≤0.01, ***P≤0.001. BMSCs, bone
mesenchymal stromal cells; CML, chronic myelogenous leukemia; HD,
healthy donor; CFU-F, colony forming units of fibroblast; CFU-GM,
colony forming units of granulocyte/monocyte; CFU-E, colony forming
units of erythroid; BFU-E, burst forming units of erythroid. |
Increased cellular senescence of
CML-BMSCs
The current study identified that BMSCs gradually
changed from a fibroblast-like shape into a large and irregular
shape, which was previously described as a typical senescence
characteristic (21). In view of this
finding, SA-β-gal stain was used to assess the percentage of BMSCs
undergoing senescence. As demonstrated in Fig. 2C, the percentage of senescent HD-BMSC
(22.33±1.61%) was significantly decreased compared with the
percentage of senescent CML-BMSC (60.17±2.66%).
Diminished hematopoietic support
capacity of CML-BMSCs
An LTC-IC assay was implemented to evaluate whether
the hematopoietic support capacity of CML-BMSCs was impaired
compared with that of HD-BMSCs. By counting the number of CFUs, the
current study identified that the LTC-IC frequency (the total
number of CFU-GMs, CFU-Es and BFU-Es) was 2.6-fold higher following
co-culture with HD-BMSCs compared with co-culture with CML-BMSCs,
as well as bigger size of CFU (Fig.
2D upper panel). In detail, CD34+ HCs cultured with
CML-BMSCs demonstrated decreased numbers of CFU-GM (103.25±2.87 vs.
267.25±67.54), CFU-E (15.25±6.24 vs. 45.00±3.92) and BFU-E
(5.50±2.65 vs. 29.25±12.31) compared with those cultured with
HD-BMSCs (Fig. 2D lower panel). The
current results identified that CML-BMSCs exhibited a less
effective hematopoietic support capability compared with
HD-BMSCs.
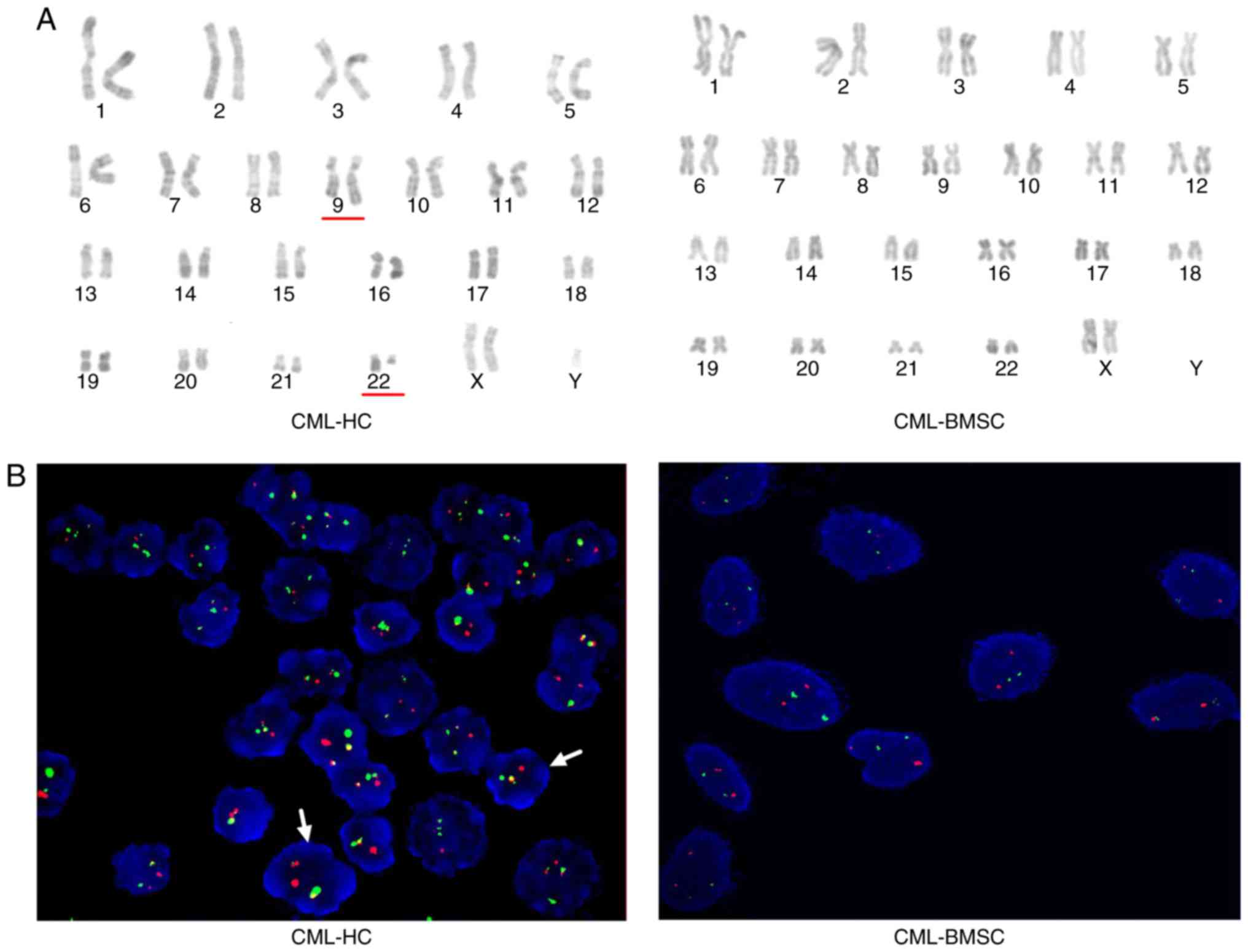
Absence of BCR-ABL in CML-BMSCs
To gain further insight into CML-BMSCs, CC analysis
and FISH analysis were used to explore cytogenetic alterations in
BMSCs and HCs from 15 patients with CML and 6 HDs. The current
study revealed that the Ph chromosome could be identified in all
CML-HCs that were analyzed by CC. The FISH results demonstrated
that all CML-HCs as indicated by the yellow hybridization signal.
However, neither no chromosomal aberrations were identified in
CML-BMSCs using CC analysis (Fig.
3A), no yellow signal was detected by FISH analysis (Fig. 3B and Table
II). As expected, HCs and BMSCs in all HDs exhibited a normal
karyotype (data not shown).
 | Table II.Chromosomal aberrations in BMSCs. |
Table II.
Chromosomal aberrations in BMSCs.
|
| Karyotype | BCR/ABL dual
fusion |
|---|
|
|
|
|
|---|
| Patient no. | HCs | BMSCs | HCs | BMSCs |
|---|
| 1 |
46,XY,t(9;22)(q34;q11.2)[16] | 46,XY[20] | + | − |
| 2 |
46,XY,t(9;22)(q34;q11.2)[12] | 46,XY[14] | + | − |
| 3 |
46,XX,t(9;22)(q34;q11.2)[20] | 46,XX[17] | + | − |
| 4 |
46,XY,t(9;22)(q34;q11.2)[13] | 46,XY[13] | + | − |
| 5 |
46,XY,t(9;22)(q34;q11.2)[11] | 46,XY[20] | + | − |
| 6 |
46,XY,t(9;22)(q34;q11.2)[17] | 46,XY[20] | + | − |
| 7 |
46,XX,t(9;22)(q34;q11.2)[19] | 46,XX[20] | + | − |
| 8 |
46,XY,t(9;22)(q34;q11.2)[15] | 46,XY[18] | + | − |
| 9 |
46,XX,t(9;22)(q34;q11.2)[18] | 46,XX[12] | + | − |
| 10 |
46,XY,t(9;22)(q34;q11.2)[12] | 46,XY[20] | + | − |
| 11 |
46,XX,t(9;22)(q34;q11.2)[20] | 46,XX[13] | + | − |
| 12 |
46,XX,t(9;22)(q34;q11.2)[14] | 46,XX[13] | + | − |
| 13 |
44,XX,t(9;22)(q34;q11.2)[16] | 46,XX[20] | + | − |
| 14 |
46,XY,t(9;22)(q34;q11.2)[15] | 46,XY[16] | + | − |
| 15 |
46,XX,t(9;22)(q34;q11.2)[20] | 46,XX[14] | + | − |
Discussion
Previously, several studies have proposed a
hypothesis that functional and genetic alterations of stromal cells
would induce myeloid malignancies (22,23).
Therefore, the current study focused on BMSCs to explore whether
their functional and genetic characteristics were altered in
CML-BMM.
The current study identified that there were no
significant differences in surface marker expression and the
ability of multipotential differentiation between the adherent
cells derived from HDs and patients with CML. Adherent cells from
both HDs and patients with CML conformed to the minimal definition
criteria for BMSCs proposed by the ISCT. In addition, the
separation method used in the current study was confirmed to be
reliable. The current study revealed that the growth rate of
CML-BMSCs was impaired compared with that of HD-BMSCs. Previous
studies have reported that impaired proliferation is not associated
with cell cycle arrest or increased rates of apoptosis (20), but is associated with an increased
rate of aging (14). The current
study identified that CML-BMSCs were more susceptible to cellular
senescence compared with HD-BMSCs. The promyelocytic leukemia gene
plays a key role in the progress of BMSCs senescence (24). Cellular senescence in BMSCs may affect
their hematopoietic support capacity (25). The current study demonstrated that the
frequency of LTC-ICs following co-culture with HD-BMSCs was
2.6-fold higher compared with that of CML-BMSCs, indicating that
CML-BMSCs exhibited an impaired hematopoietic support capacity
compared with HD-BMSCs. Within the BMM in hematologic neoplasms,
malignant HCs and healthy HCs are affected by exosomes, microRNAs,
long non-coding RNAs and cytokines secreted by stromal cells
(26–28). Among them, to the best of our
knowledge, the research seem to focus more on the effects of
cytokines on hematopoietic support capacity in BMM at present
(3,29). A limitation of the current study is
that only the phenomenon of biological characteristics of CML-BMSCs
were provided, while in-depth research on the mechanism of the
impaired biological characteristics was not conducted. In the near
future, the effects of cytokine changes on the impaired
hematopoietic support capacity of CML-BMSCs may be investigated. It
has previously been reported that malignant HCs may remodel and
transform a healthy BMM into an inflammatory environment (30,31). When
exposed to an inflammatory environment in vivo, the
epigenetics of BMSCs would be altered and functions of BMSCs would
be impaired, including their hematopoietic support capacity
(12,13). A previous study demonstrated that the
capacity of BMSCs to support healthy HCs was impaired, however they
selectively promoted the proliferation of malignant HCs (32).
Growing evidence has demonstrated that genetic
abnormalities exist in BMSCs derived from leukemia patients
(15,33). This suggests that a primary BMSC
defect either causes or supports hematologic malignancy. This
hypothesis challenges the popular theory that hematological
malignancies originate exclusively from intrinsic genetic defects
in HCs (34). The current study did
not detect any of the cytogenetic abnormalities in CML-BMSCs that
have been identified in previous studies (35,36). As
previously mentioned, the current study has verified that the
biological function of BMSCs was impaired when derived from
patients with CML patients compared with that of HD-BMSCs.
Functional inhibition has been demonstrated to be associated with
DNA methylation in AML and MDS-derived BMSCs (12,13).
Abnormalities of DNA methylation may lead to gene instability
(37,38). Therefore, the current study assumed
that BMSCs with altered biological functions may exhibit a
potential risk of gene mutation. The accumulation of genetic
mutations may eventually lead to chromosomal aberrations (34,39). BMSCs
and HCs may similarly be affected by a predisposing genetic defect
and therefore contribute equally to the manifestation of CML
(40).
Whether genetic stability of BMSCs could be
maintained in vitro is of note. A previous study has
revealed genetic material of BMSCs remains intact up to and
including passage 9 (41). All BMSCs
adopted in the current study for chromosomal analysis were within
passage 5. Therefore, it is likely the BMSCs used in the current
study only acquired additional mutations at a very low
frequency.
In summary, the current study performed a
comprehensive analysis that demonstrated the function of CML-BMSCs
was inhibited but no cytogenetic abnormalities were detected. The
current study has provided further insight into the pathophysiology
of the CML-BMM with the view of providing an improved understanding
of BMSCs.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Fujian
Medical University Professor Fund (grant no. JS14024) and Fujian
Provincial Natural Fund (grant no. 2015J01472).
Availability of data and materials
The analyzed data are available from the
corresponding author upon reasonable request.
Authors' contributions
JQX and HFH designed the experiments. JQX and JDC
performed the laboratory work for this study. BW and XCH recruited
the patients. BW participated in data analysis. XCH performed part
of the laboratory work and drafted the corresponding part of the
manuscript. JQX and HFH worked on the manuscript. All the authors
have read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The Ethical Committee of Fujian Medical University
Union Hospital (approval no. 2016KY053). Participants provided
written informed consent for their inclusion in the present
study.
Patient consent for publication
All patients provided written informed consent for
the publication of their data and any associated images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Apperley JF: Chronic myeloid leukaemia.
Lancet. 385:1447–1459. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bruns I, Czibere A, Fischer JC, Roels F,
Cadeddu RP, Buest S, Bruennert D, Huenerlituerkoglu AN, Stoecklein
NH, Singh R, et al: The hematopoietic stem cell in chronic phase
CML is characterized by a transcriptional profile resembling normal
myeloid progenitor cells and reflecting loss of quiescence.
Leukemia. 23:892–899. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang B, Ho YW, Huang Q, Maeda T, Lin A,
Lee SU, Hair A, Holyoake TL, Huettner C and Bhatia R: Altered
microenvironmental regulation of leukemic and normal stem cells in
chronic myelogenous leukemia. Cancer Cell. 21:577–592. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Penserga ET and Skorski T: Fusion tyrosine
kinases: A result and cause of genomic instability. Oncogene.
26:11–20. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Melo JV and Barnes DJ: Chronic myeloid
leukaemia as a model of disease evolution in human cancer. Nat Rev
Cancer. 7:441–453. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Goldman JM and Melo JV: Targeting the
BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med.
344:1084–1086. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Goldman JM and Melo JV: Chronic myeloid
leukemia-advances in biology and new approaches to treatment. N
Engl J Med. 349:1451–1464. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou H, Mak PY, Mu H, Mak DH, Zeng Z,
Cortes J, Liu Q, Andreeff M and Carter BZ: Combined inhibition of
β-catenin and Bcr-Abl synergistically targets tyrosine kinase
inhibitor-resistant blast crisis chronic myeloid leukemia blasts
and progenitors in vitro and in vivo. Leukemia. 31:2065–2074. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jin L, Tabe Y, Konoplev S, Xu Y, Leysath
CE, Lu H, Kimura S, Ohsaka A, Rios MB, Calvert L, et al: CXCR4
up-regulation by imatinib induces chronic myelogenous leukemia
(CML) cell migration to bone marrow stroma and promotes survival of
quiescent CML cells. Mol Cancer Ther. 7:48–58. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schroeder T, Geyh S, Germing U and Hass R:
Mesenchymal stromal cells in myeloid malignancies. Blood Res.
51:225–232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Frenette PS, Pinho S, Lucas D and
Scheiermann C: Mesenchymal stem cell: Keystone of the hematopoietic
stem cell niche and a stepping-stone for regenerative medicine.
Annu Rev Immunol. 31:285–316. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Geyh S, Rodriguez-Paredes M, Jäger P,
Khandanpour C, Cadeddu RP, Gutekunst J, Wilk CM, Fenk R, Zilkens C,
Hermsen D, et al: Functional inhibition of mesenchymal stromal
cells in acute myeloid leukemia. Leukemia. 30:683–691. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Geyh S, Oz S, Cadeddu RP, Fröbel J,
Brückner B, Kündgen A, Fenk R, Bruns I, Zilkens C, Hermsen D, et
al: Insufficient stromal support in MDS results from molecular and
functional deficits of mesenchymal stromal cells. Leukemia.
27:1841–1851. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao Y, Wu D, Fei C, Guo J, Gu S, Zhu Y,
Xu F, Zhang Z, Wu L, Li X and Chang C: Down-regulation of Dicer1
promotes cellular senescence and decreases the differentiation and
stem cell-supporting capacities of mesenchymal stromal cells in
patients with myelodysplastic syndrome. Haematologica. 100:194–204.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Blau O, Baldus CD, Hofmann WK, Thiel G,
Nolte F, Burmeister T, Türkmen S, Benlasfer O, Schümann E, Sindram
A, et al: Mesenchymal stromal cells of myelodysplastic syndrome and
acute myeloid leukemia patients have distinct genetic abnormalities
compared with leukemic blasts. Blood. 118:5583–5592. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arber DA, Orazi A, Hasserjian R, Thiele J,
Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M and Vardiman JW:
The 2016 revision to the World Health Organization classification
of myeloid neoplasms and acute leukemia. Blood. 127:2391–2405.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bernheim A: Cytogenomics of cancers: From
chromosome to sequence. Mol Oncol. 4:309–322. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Simons A, Shaffer LG and Hastings RJ:
Cytogenetic nomenclature: Changes in the ISCN 2013 compared to the
2009 Edition. Cytogenet Genome Res. 141:1–6. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dominici M, Le Blanc K, Mueller I,
Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A,
Prockop Dj and Horwitz E: Minimal criteria for defining multipotent
mesenchymal stromal cells. The International Society for cellular
therapy position statement. Cytotherapy. 8:315–317. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lopez-Villar O, Garcia JL, Sanchez-Guijo
FM, Robledo C, Villaron EM, Hernández-Campo P, Lopez-Holgado N,
Diez-Campelo M, Barbado MV, Perez-Simon JA, et al: Both expanded
and uncultured mesenchymal stem cells from MDS patients are
genomically abnormal, showing a specific genetic profile for the
5q-syndrome. Leukemia. 23:664–672. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Campisi J and d'Adda di Fagagna F:
Cellular senescence: When bad things happen to good cells. Nat Rev
Mol Cell Biol. 8:729–740. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rupec RA, Jundt F, Rebholz B, Eckelt B,
Weindl G, Herzinger T, Flaig MJ, Moosmann S, Plewig G, Dörken B, et
al: Stroma-mediated dysregulation of myelopoiesis in mice lacking I
kappa B alpha. Immunity. 22:479–491. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schepers K, Pietras E, Reynaud D, Flach J,
Binnewies M, Garg T, Wagers AJ, Hsiao EC and Passegué E:
Myeloproliferative neoplasia remodels the endosteal bone marrow
niche into a self-reinforcing leukemic niche. Cell Stem Cell.
13:285–299. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fu S, Wei J, Wang G, Wang B, Wang Y, Lai X
and Huang H: The key role of PML in IFN-α induced cellular
senescence of human mesenchymal stromal cells. Int J Oncol.
46:351–359. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Walenda T, Bork S, Horn P, Wein F,
Saffrich R, Diehlmann A, Eckstein V, Ho AD and Wagner W: Co-culture
with mesenchymal stromal cells increases proliferation and
maintenance of haematopoietic progenitor cells. J Cell Mol Med.
14:337–350. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kumar B, Garcia M, Weng L, Jung X,
Murakami JL, Hu X, McDonald T, Lin A, Kumar AR, DiGiusto DL, et al:
Acute myeloid leukemia transforms the bone marrow niche into a
leukemia-permissive microenvironment through exosome secretion.
Leukemia. 32:575–587. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barrera-Ramirez J, Lavoie JR, Maganti HB,
Stanford WL, Ito C, Sabloff M, Brand M, Rosu-Myles M, Le Y and
Allan DS: Micro-RNA profiling of exosomes from marrow-derived
mesenchymal stromal cells in patients with acute myeloid leukemia:
Implications in leukemogenesis. Stem Cell Rev. 13:817–825. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shah MY, Ferracin M, Pileczki V, Chen B,
Redis R, Fabris L, Zhang X, Ivan C, Shimizu M, Rodriguez-Aguayo C,
et al: Cancer-associated rs6983267 SNP and its accompanying long
noncoding RNA CCAT2 induce myeloid malignancies via unique
SNP-specific RNA mutations. Genome Res. 28:432–447. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hong DS, Angelo LS and Kurzrock R:
Interleukin-6 and its receptor in cancer: Implications for
translational therapeutics. Cancer. 110:1911–1928. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hoggatt J, Kfoury Y and Scadden DT:
Hematopoietic stem cell niche in health and disease. Annu Rev
Pathol. 11:555–581. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schepers K, Campbell TB and Passegué E:
Normal and leukemic stem cell niches: Insights and therapeutic
opportunities. Cell Stem Cell. 16:254–267. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hanoun M, Zhang D, Mizoguchi T, Pinho S,
Pierce H, Kunisaki Y, Lacombe J, Armstrong SA, Dührsen U and
Frenette PS: Acute myelogenous leukemia-induced sympathetic
neuropathy promotes malignancy in an altered hematopoietic stem
cell niche. Cell Stem Cell. 15:365–375. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jurczyszyn A, Czepiel J, Gdula-Argasińska
J, Perucki W, Skotnicki AB and Majka M: The analysis of the
relationship between multiple myeloma cells and their
microenvironment. J Cancer. 6:160–168. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sperling AS, Gibson CJ and Ebert BL: The
genetics of myelodysplastic syndrome: From clonal haematopoiesis to
secondary leukaemia. Nat Rev Cancer. 17:5–19. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jootar S, Pornprasertsud N, Petvises S,
Rerkamnuaychoke B, Disthabanchong S, Pakakasama S, Ungkanont A and
Hongeng S: Bone marrow derived mesenchymal stem cells from chronic
myeloid leukemia t(9;22) patients are devoid of Philadelphia
chromosome and support cord blood stem cell expansion. Leuk Res.
30:1493–1498. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wöhrer S, Rabitsch W, Shehata M, Kondo R,
Esterbauer H, Streubel B, Sillaber C, Raderer M, Jaeger U,
Zielinski C and Valent P: Mesenchymal stem cells in patients with
chronic myelogenous leukaemia or bi-phenotypic Ph+ acute leukaemia
are not related to the leukaemic clone. Anticancer Res.
27:3837–3841. 2007.PubMed/NCBI
|
|
37
|
Hamidi T, Singh AK and Chen T: Genetic
alterations of DNA methylation machinery in human diseases.
Epigenomics. 7:247–265. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Akhavan-Niaki H and Samadani AA: DNA
methylation and cancer development: Molecular mechanism. Cell
Biochem Biophys. 67:501–513. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Weckselblatt B and Rudd MK: Human
structural variation: Mechanisms of chromosome rearrangements.
Trends Genet. 31:587–599. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
University of Chicago Hematopoietic
Malignancies Cancer Risk Team: How I diagnose and manage
individuals at risk for inherited myeloid malignancies. Blood.
128:1800–1813. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ben-David U, Mayshar Y and Benvenisty N:
Large-scale analysis reveals acquisition of lineage-specific
chromosomal aberrations in human adult stem cells. Cell Stem Cell.
9:97–102. 2011. View Article : Google Scholar : PubMed/NCBI
|