Introduction
Glioma, a rapidly progressive malignant brain tumor,
is the most common and aggressive type of primary brain neoplasms
found in adults, leading to growth of the neural microenvironment,
particularly in the blood vessels (1–3). Owing to
its high invasiveness, patients with glioma invariably cannot
undergo surgery and when they do it is easy to relapse, resulting
in short survival (2,4). Glioma patients have a poor outcome with
a 5-year survival rate of 9.8%, despite multimodal treatment
options including surgery, radiation and chemotherapy (5).
MicroRNAs (miRNAs), small non-coding regulatory
molecules, bind to the 3′-untranslated region (3′-UTR) of target
mRNAs to degrade mRNA and/or inhibit protein translation at
post-transcription (6,7). Increasing evidence suggests that miRNAs
mediate the carcinogenic process with regard to cell proliferation,
invasion and apoptosis (8,9). miR-153, a novel tumor-associated miRNA
is downregulated in various types of cancer, including non-small
cell lung cancer, gastric cancer, breast cancer, pancreatic ductal
adenocarcinoma and melanoma (10–16).
miR-153 suppresses gastric cell migration and invasion by
inhibiting SNAI1-induced EMT (10).
miR-153 decreased the ability of migration and invasion by
targeting ADAM19 in non-small cell lung cancer cells (15). A low expression of miR-153 promoted
breast cancer cell apoptosis via targeting HECTD3 (13). Zeng et al observed that
microRNA-153 inhibited melanoma cell proliferative and invasive
abilities by targeting SNAI1 and inverse correlations between
miR-153 and SNAI1 (14). However,
miR-153 was found to be upregulated and promoted development and
progression in colorectal and prostate cancer (15,17). Thus,
the roles of miR-153 seem to be different, depending on cancer
types. However, the precise mechanism of miR-153 in glioma has not
been investigated.
SNAI1 (snail) acts as transcription repressor and is
a member of the zinc finger superfamily. SNAI1 shares conserved
SNAG domain with other members at N-terminal, whose function is
mediating transcription repression (18). SNAI1 is a candidate marker of
epithelial-mesenchymal transition (EMT), which endows normal cells
with the ability to metastasize by repressing the expression of
epithelial cell marker E-cadherin (19). It has been reported that esophageal
squamous cell carcinoma patients with SNAI1 high expression had
worse prognosis than those with low expression of SNAI1 (20). Dong et al have reported that
SNAI1 was upregulated in glioma and interference of SNAI1 could
inhibit glioma cell proliferation and migration (21). However, the clinical significance of
Snail in glioma remains to be determined.
In the present study, we investigated miR-153 low
expression in glioma cancer tissues and cells. The results showed
that miR-153 suppressed glioma cell invasion through targeting
SNAI1 and SNAI1 revealed the partial function of miR-153. In
addition, miR-153 low expression was correlated with poor
prognostic parameters in glioma.
Materials and methods
Patients and clinical tissue
specimens
Paired cancer tissues and corresponding
paracancerous tissues were obtained from 55 patients who underwent
glioma in the Yantai Yuhuangding Hospital (Yantai, China) between
January 2015 and July 2017, and were stored at −80°C before
analysis. None of the patients had received preoperative chemo- or
radiotherapy prior to surgery. Of this cohort, patients diagnosed
at early stage (I/II) were 27, while the remaining 28 patients were
at an advanced stage (III/V) (Table
I). Written informed consent was obtained from patients and the
study was approved by the Ethical Committee of Yantai Yuhuangding
Hospital.
 | Table I.miR-153 expression and
clinicopathological features in 55 paired gliomas. |
Table I.
miR-153 expression and
clinicopathological features in 55 paired gliomas.
|
|
| miR-153
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Cases (n=55) | 27 High (%) | 28 Low (%) | P-valuea |
|---|
| Age (years) |
|
|
| 0.022a |
|
<50 | 26 | 17 (53.8) | 9
(34.6) |
|
| ≥50 | 29 | 10 (34.5) | 19 (65.5) |
|
| Sex |
| Male | 28 | 15 (53.6) | 13 (46.4) | 0.498 |
|
Female | 27 | 12 (44.4) | 15 (55.6) |
|
| Tumor size (mm) |
|
|
| 0.052 |
| ≤4.0 | 28 | 17 (68.0) | 11 (32.0) |
|
|
>4.0 | 27 | 10 (33.3) | 17 (66.7) |
|
| TNM stage |
|
|
| 0.043a |
|
I–II | 25 | 16 (64.0) | 9
(36.0) |
|
|
III–IV | 30 | 11 (36.7) | 19 (63.3) |
|
| Lymph node
metastasis |
|
|
| 0.022a |
| No | 24 | 16 (66.7) | 8
(33.3) |
|
|
Yes | 31 | 11 (35.5) | 20 (64.5) |
|
| SNAI1 |
|
|
| 0.042a |
|
Negative | 29 | 18 (62.1) | 11 (37.9) |
|
|
Positive | 26 | 9
(34.6) | 17 (65.4) |
|
Cell lines and culture condition
Normal immortalized gliocyte HEB cells and the
glioma cell lines U-87MG ATCC and U251 were purchased from American
Type Culture Collection (cat. no. HTB-14™; ATCC, Rockville, MD,
USA). All the cells were cultured in RPMI-1640 medium containing
10% fetal bovine serum (FBS) (both from Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) at 37°C with 5% CO2.
U-87MG ATCC cells were of CNS origin and are likely to be a
bonafide human glioblastoma cell line considering their mRNA
expression profile. Thus, U-87MG ATCC cells can be used in glioma
research and are distinct from U-87MG ATCC Uppsala cells
established in 1968 at the University of Uppsala (22).
Transfection
Plasmids including pcDNA3.1-SNAI1, as well as
miR-153 mimic and inhibitor together with their negative control
were purchased from GenePharma (Shanghai, China). Cells were seeded
in 6-well plates with a density of 3×105 and cultivated
at 37°C overnight. Lipofectamine 2000™ (Invitrogen; Thermo Fisher
Scientific) was to perform the transfection in U-87MG ATCC and U251
cells, as per the manufacturer's protocol.
RNA isolation and reverse
transcription quantitative-PCR (RT-qPCR)
Total RNAs and total miRNAs were extracted from
glioma tissues and cells using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific) and mirVana™ miRNA Isolation kit (Ambion,
Austin, TX, USA). RNAs were reverse transcribed to produce first
cDNA using PrimeScript™ II 1st Strand cDNA Synthesis kit or TaqMan
MicroRNA reverse Transcription kit (Takara, Dalian, China). Next,
TaqMan microRNA assays and the TaqMan Gene Expression assays
(Applied Biosystems; Thermo Fisher Scientific, Inc.) were applied
to carried out quantitative PCR on the Step-one plus real-time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
relative expression of mRNAs and miRNAs was calculated with
2−∆∆Cq method with GAPDH or U6 which were normalized,
respectively (23).
Transwell assay
Transwell assay was employed to perform invasive
activity with Matrigel (Clontech, Mountain View, CA, USA) added in
the Transwell chamber (8.0 µm pore size; Corning Incorporated,
Corning, NY, USA). Before test, we suspended glioma cells
(1×105) using RPMI-1640 medium, and placed the Transwell
chamber into 24-well plate. Cells (200 µl) were then suspensded in
the upper chamber, followed by the addition of 500 µl medium
containing 15% FBS in the lower chamber and incubation at 37°C for
24 h. The cells that adhered to the lower surface were stained for
about 30 min at 37°C using 0.5% crystal violet. The invasive cells
were captured using a microscope (BX51 Olympus; Olympus
Corporation, Tokyo, Japan) and counted at five random fields.
Protein extraction and western blot
analysis
Glioma cells were lysed in RIPA lysis buffer on ice
for 30 min and after centrifugation at at 12,000 × g for 15 min at
4°C, the supernatant was removed. Protein concentration was
quantified using Bradford Protein assay kit (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). Equal amounts of total protein (50 µg)
were separated using 10% SDS-PAGE. After electrophoresis, the blots
were transferred onto PVDF membranes (Bio-Rad) and with 5% skim
milk powder. Subsequently, the membranes were incubated at 4°C
overnight with anti-SNAI1 mouse monoclonal primary antibody (cat.
no. ab167609; 1:1,000; Abcam, Cambridge, UK) and GAPDH (cat. no.
G5262; 1:3,000; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
acted as internal reference. Subsequently, the membrane was
incubated using mouse IgG horseradish peroxidase-conjugated
secondary antibody (cat. no. sc-2357; 1:4,000; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) and visualized using the
enhanced chemiluminescence kit (ECL; EMD Millipore, Billerica, MA,
USA). Densitometry was calculated using Bio-Rad Gel Doc XR
instrument (Bio-Rad).
Plasmid construction and luciferase
reporter assay
TargetScan (http://www.targetscan.org/vert_71/) was employed to
predict target genes of miR-153 and the binding site was also
predicted. The 3′UTR fragment of the target gene, containing the
binding site of miR-153, was inserted into the pmirGLO Vector (WT;
Promega, Madison, WI, USA). A mutant (MUT) of 3′-untranslated
region (3′UTR) of the target gene was employed using QuikChange
Multi Site-Directed Mutagenesis kit (Stratagene; Agilent
Technologies, Inc., Santa Clara, CA, USA), according to the
manufacturer's protocol. Then, miR-153 was cloned into the
previously stored pmirGlo vector. We co-transfected recombinant
reporter plasmids WT or MUT and either the pmirGlo-miR-153 or the
miR-scramble control into U-87MG ATCC and U251 cells. After 48 h,
the Dual Luciferase Reporter assay system (Promega) was utilized to
measure the luciferase activities.
Statistical analysis
Student's t-test or ANOVA and Scheffe test were used
to perform the statistical analysis. Pearson χ2 test was
used to test the correlation between miR-153 and SNAI1 and
clinicopathological characteristics of gastric carcinoma. Survival
curves were plotted using standard Kaplan-Meier and log-rank tests.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of miR-153 and SNAI1 is
inversely correlated in glioma
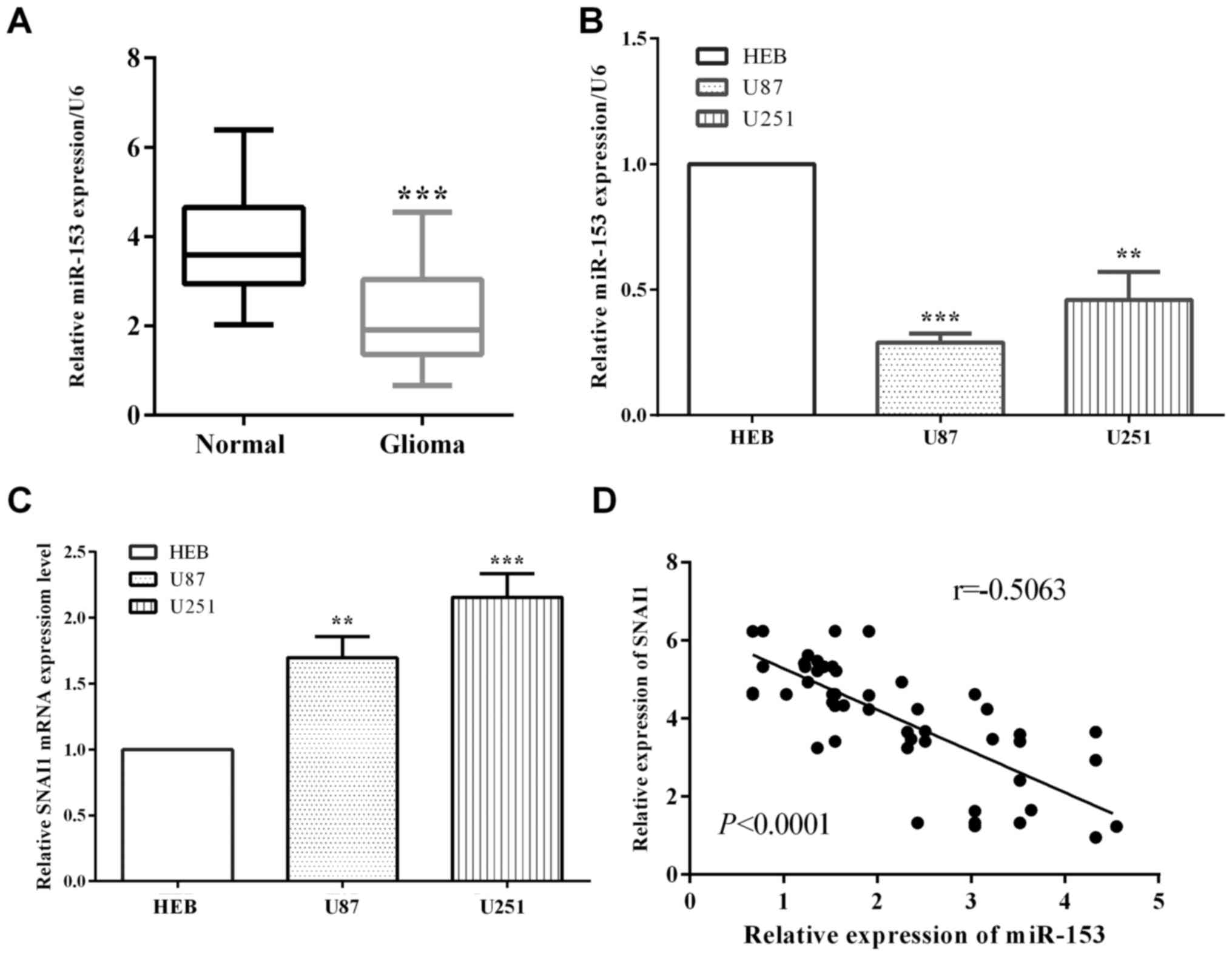
We assessed miR-153 level in glioma and
corresponding paracancerous tissues, as well as two glioma cell
lines (U-87MG ATCC and U251) and human normal immortalized gliocyte
HEB cells. As expected, the relative expression of miR-153 in
glioma tissues was lower than the corresponding paracancerous
tissues (P<0.0001) (Fig. 1A).
Compared with normal immortalized gliocyte HEB cells, miR-153 is
downregulated in glioma cell linesU-87MG ATCC (P<0.0001) and
U251 (P=0.0010) (Fig. 1B). On the
other hand, SNAI1 is overexpressed in glioma cell lines U-87MG ATCC
(P=0.0017) and U251 (P=0.0004) (Fig.
1C). Additionally, our results of miR-153 and SNAI1 expression
in 55 glioma patients indicated an inverse correlation between
miR-153 and SNAI1 in glioma tissue specimens (P<0.0001,
r=−0.7469) (Fig. 1D).
miR-153 low expression suppressed
glioma invasion
To investigate the function of miR-153 on the
invasion of glioma cells, miR-153 was upregulated/downregulated in
U-87MG ATCC (P=0.0026 and 0.0390) and U251 (P=0.0040 and 0.0093)
cell line through transfected miR-153 mimic/miR-153 inhibitor
(Fig. 2A). Transwell assays were
employed to detect the influence of altering miR-153 levels on
invasion. We found that the cell invasive number was significantly
decreased with overexpression of miR-153 in U-87MG ATCC (P=0.0161)
and U251 (P=0.0233) cells. On the contrary, low expression of
miR-153 significantly promoted cell invasive ability of U-87MG ATCC
(P=0.0144) and U251 (P=0.0011) (Fig.
2B).
miR-153 directly targeted SNAI1 and
regulated SNAI1 expression
We have previously demonstrated that miR-153
affected cell invasion, however the regulation of miR-153 rmains to
be determined. We identified SNAI1 was a putative binding site of
miR-153 by TargetScan algorithms (http://www.targetscan.org/vert_71/). To confirm that
SNAI1 is a direct target of miR-153, we mutated the binding region
from CUAUGCAA to CGAGGAAA and inserted it into the pmirGlo vector
(pmirGlo-SNAI1-WT or pmirGlo-SNAI1-MUT) (Fig. 3A). We co-transfected pmirGlo-SNAI1-WT
or pmirGlo-SNAI1-MUT and miR-153 mimic or control into U-87MG ATCC
and U251 cells. The luciferase reporter assays showed that
co-transfection with miR-153 mimic and pmirGlo-SNAI1-WT prominently
suppressed luciferase activity compared with co-transfected control
mimic and pmirGlo-SNAI1-WT in both U-87MG ATCC (P=0.0011) and U251
(P=0.0008) cells. On the other hand, there was almost no difference
between co-transfection with pmirGlo-SNAI1-MUT and miR-153 mimic
and control in U-87MG ATCC (P=0.7854) and U251 (P=0.5161) cells
(Fig. 3B). These results suggested
that miR-153 can directly target and suppress the SNAI1 expression
in U-87MG ATCC and U251 cells.
SNAI1 reversed the partial impact of
miR-153
To classify whether miR-153 suppressed invasive
activity by regulating SNAI1 expression, we co-transfected miR-153
and SNAI1. As expected, when we transfected miR-153 mimic to
overexpressed miR-153, the expression of SNAI1 was decreased both
at the mRNA and protein level in U-87MG ATCC (P=0.0013) and U251
(P=0.0016) cells. SNAI1 expression was reduced when transfected
with miR-153 mimic (P=0.0342 and 0.0220), which was re-expressed
via transfected SNAI1 (Fig. 4A).
SNAI1 overexpression partially attenuated the inhibiting effect of
miR-153 on cell invasion, suggesting that SNAI1 is involved in
miR-153 mediated biological function in U-87MG ATCC (P=0.0117) and
U251 (P=0.0210) (Fig. 4B). Therefore,
SNAI1 reversed the partial function of miR-153.
miR-153 low expressed predicted poor
prognosis
According to miR-153 expression level, 55 gastric
cancer patients were divided into the high expression group
[miR-153(+)] and low expression group [miR-153(−)], with 27 and 28
patients, respectively. Detailed information on the 55 patients
including factors such as sex, age, tumor size, TNM stage, lymph
node metastasis and SNAI1 are presented in Table I. The results showed that the
expression of miR-153 was dependent on age (P=0.022), TNM stage
(P=0.043), lymph node metastasis (P=0.022) and SNAI1 (P=0.042),
while had tendency association with tumor size (P=0.052). However,
there was no correlation between miR-153 with sex (P=0.498).
In addition, Kaplan-Meier survival analysis of the
overall survival (OS) and disease-free survival (DFS) revealed that
the OS and DFS in miR-153(+) group was significantly higher than
miR-153(−) group (log-rank P=0.0247 and 0.0107) (Fig. 4C).
Discussion
Glioma is a kind of brain intrinsic tumor and
limited progression concerning control thereof has been made in the
past 30 years (1). High-grade glioma
is among the most aggressive types of malignancy, for which there
is currently no cure; therefore, finding new biomarkers for glioma
is crucial (24). MicroRNAs typically
block translation or degrade target messenger RNAs (mRNAs) to
inhibit target gene expression at the post-transcriptional level
(25–28). miR-153 was reported to be
downregulated in glioma and inhibit mice tumor growth and promote
ability of clone formation (24). In
the present study, we determined that SNAI1 was a direct target of
miR-153 in glioma cell lines U-87MG ATCC and U251. Furthermore, low
expression of miR-153 in glioma tissues and cell lines was
identified, while SNAI1 was lowly expressed versus corresponding
paracancerous tissues and the normal immortalized gliocyte cells.
In consideration of these results, we strongly believe that the
impact of microRNA-153 on invasion may be through direct inhibition
of SNAI1. To the best of our knowledge, this is the first time
miR-153 has been suggested to directly target SNAI1 and affect
invasion through regulation of SNAI1 in glioma. Additionally, this
is the first study to examine the role of miR-153 on survival of
patients with glioma.
The function of miR-153 on cell proliferation,
migration, invasion and apoptosis has been previously studied in
most cancer cells (12,14–16,24). Bai
et al have shown that miR-153 acts as a prognostic marker
and inhibits the migratory and invasive ability of human pancreatic
ductal adenocarcinoma cells by targeting SNAI1 (14). Similar findings were reported by Niu
et al who identified that miR-153 inhibited cell
proliferation and invasion in osteosarcoma cells by targeting
TGF-β2 (29). Similarly, our results
demonstrated that miR-153 suppressed invasive capacity via
targeting SNAI1 in glioma cells U-87MG ATCC and U251. Furthermore,
miR-153 low expression can predict poor prognosis, and to the best
of our knowledge, this is the first study to identify miR-153 is
associated with the prognosis of patients with glioma.
In our study, SNAI1 was related to the impact of
miR-153 on cell invasion in glioma cells U-87MG ATCC and U251.
SNAI1 is a promoting factor of EMT, which endows normal cells with
the ability to metastasize by repressing expression of E-cadherin
(19). Furthermore, overexpressed
SNAI1 has been reported in esophageal squamous cell carcinoma,
pancreatic ductal adenocarcinoma and melanoma (12,14,16). It
has been reported that patients with high expression of SNAI1 had
worse prognosis than those with low expression in esophageal
squamous cell carcinoma (20). Dong
et al have reported that SNAI1 was upregulated in glioma and
interference of SNAI1 could inhibit glioma cell proliferation and
migration (21). In our study, SNAI1
was overexpressed in glioma tissues and cell lines U-87MG ATCC and
U251 versus paracancerous tissues and immortalized gliocyte HEB
cells. Therefore, transfection of miR-153 mimic could inhibit SNAI1
expression, thereby causing cell invasion. In addition, SNAI1 was
downregulated by miR-153 and had a negative association with the
expression of miR-153 (12,14,16).
Consistent with all the findings, we suggest that SNAI1 is a direct
target of miR-153 and is mediated by miR-153 in glioma cell
invasion.
In conclusion, the present findings demonstrate that
miR-153 could influence cell invasion by mediating SNAI1 expression
in glioma cells. miR-153 may act as a prognostic marker to predict
survival of glioma patients.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
WZ and CYY contributed to the conception of the
study. JJ contributed significantly to the data analysis and study
preparation. WK and HX performed the data analyses and wrote the
study. HZ helped perform the analysis with constructive
discussions. All authors have read and approved the final
study.
Ethics approval and consent to
participate
Written informed consent was obtained from patients
and the study was approved by the Ethical Committee of Yantai
Yuhuangding Hospital (Yantai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Westphal M and Lamszus K: The neurobiology
of gliomas: From cell biology to the development of therapeutic
approaches. Nat Rev Neurosci. 12:495–508. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kwiatkowski SC, Guerrero PA, Hirota S,
Chen Z, Morales JE, Aghi M and McCarty JH: Neuropilin-1 modulates
TGFβ signaling to drive glioblastoma growth and recurrence after
anti-angiogenic therapy. PLoS One. 12:e01850652017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Louis DN: Molecular pathology of malignant
gliomas. Annu Rev Pathol. 1:97–117. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sayegh ET, Kaur G, Bloch O and Parsa AT:
Systematic review of protein biomarkers of invasive behavior in
glioblastoma. Mol Neurobiol. 49:1212–1244. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Munthe S, Halle B, Boldt HB, Christiansen
H, Schmidt S, Kaimal V, Xu J, Zabludoff S, Mollenhauer J, Poulsen
FR, et al: Shift of microRNA profile upon glioma cell migration
using patient-derived spheroids and serum-free conditions. J
Neurooncol. 132:45–54. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Siomi H and Siomi MC: Posttranscriptional
regulation of microRNA biogenesis in animals. Mol Cell. 38:323–332.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fabbri M: MicroRNAs and cancer: Towards a
personalized medicine. Curr Mol Med. 13:751–756. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gonzalez-Duarte RJ, Cazares-Ordonez V and
Avila-Chavez E: The microRNA biogenesis machinery: Regulation by
steroid hormones and alterations in cancer. Rev Invest Clin.
66:460–464. 2014.PubMed/NCBI
|
|
10
|
Zhang Z, Sun J, Bai Z, Li H, He S, Chen R
and Che X: MicroRNA-153 acts as a prognostic marker in gastric
cancer and its role in cell migration and invasion. Onco Targets
Ther. 8:357–364. 2015.PubMed/NCBI
|
|
11
|
Wang W, Peng B, Wang D, Ma X, Jiang D,
Zhao J and Yu L: Human tumor microRNA signatures derived from
large-scale oligonucleotide microarray datasets. Int J Cancer.
129:1624–1634. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zuo J, Wang D, Shen H, Liu F, Han J and
Zhang X: MicroRNA-153 inhibits tumor progression in esophageal
squamous cell carcinoma by targeting SNAI1. Tumour Biol.
37:16135–16140. 2016. View Article : Google Scholar
|
|
13
|
Wu X, Li L, Li Y and Liu Z: miR-153
promotes breast cancer cell apoptosis by targeting HECTD3. Am J
Cancer Res. 6:1563–1571. 2016.PubMed/NCBI
|
|
14
|
Bai Z, Sun J, Wang X, Wang H, Pei H and
Zhang Z: MicroRNA-153 is a prognostic marker and inhibits cell
migration and invasion by targeting SNAI1 in human pancreatic
ductal adenocarcinoma. Oncol Rep. 34:595–602. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shan N, Shen L, Wang J, He D and Duan C:
miR-153 inhibits migration and invasion of human non-small-cell
lung cancer by targeting ADAM19. Biochem Biophys Res Commun.
456:385–391. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zeng HF, Yan S and Wu SF: MicroRNA-153-3p
suppress cell proliferation and invasion by targeting SNAI1 in
melanoma. Biochem Biophys Res Commun. 487:140–145. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang L, Pickard K, Jenei V, Bullock MD,
Bruce A, Mitter R, Kelly G, Paraskeva C, Strefford J, Primrose J,
et al: miR-153 supports colorectal cancer progression via
pleiotropic effects that enhance invasion and chemotherapeutic
resistance. Cancer Res. 73:6435–6447. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nieto MA: The snail superfamily of
zinc-finger transcription factors. Nat Rev Mol Cell Biol.
3:155–166. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Peinado H, Ballestar E, Esteller M and
Cano A: Snail mediates E-cadherin repression by the recruitment of
the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell
Biol. 24:306–319. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Natsugoe S, Uchikado Y, Okumura H,
Matsumoto M, Setoyama T, Tamotsu K, Kita Y, Sakamoto A, Owaki T,
Ishigami S, et al: Snail plays a key role in E-cadherin-preserved
esophageal squamous cell carcinoma. Oncol Rep. 17:517–523.
2007.PubMed/NCBI
|
|
21
|
Dong Q, Cai N, Tao T, Zhang R, Yan W, Li
R, Zhang J, Luo H, Shi Y, Luan W, et al: An axis involving SNAI1,
microRNA-128 and SP1 modulates glioma progression. PLoS One.
9:e986512014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Allen M, Bjerke M, Edlund H, Nelander S
and Westermark B: Origin of the U87MG glioma cell line: Good news
and bad news. Sci Transl Med. 8:54re32016. View Article : Google Scholar
|
|
23
|
Livak KJ and Scmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cui Y, Zhao J, Yi L and Jiang Y:
microRNA-153 targets mTORC2 component rictor to inhibit glioma
cells. PLoS One. 11:e01569152016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Christodoulatos GS and Dalamaga M:
Micro-RNAs as clinical biomarkers and therapeutic targets in breast
cancer: Quo vadis? World J Clin Oncol. 5:71–81. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lauressergues D, Couzigou JM, Clemente HS,
Martinez Y, Dunand C, Bécard G and Combier JP: Primary transcripts
of microRNAs encode regulatory peptides. Nature. 520:90–93. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, maturation, target recognition and regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
28
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Niu G, Li B, Sun L and An C: MicroRNA-153
inhibits osteosarcoma cells proliferation and invasion by targeting
TGF-β2. PLoS One. 10:e01192252015. View Article : Google Scholar : PubMed/NCBI
|