Introduction
Head and neck squamous cell carcinoma (HNSCC) is one
of the most prevalent tumor types, with >500,000 newly diagnosed
cases reported globally in 2008 (1,2). Oral
squamous cell carcinoma (OSCC) is the most frequent subset of HNSCC
worldwide (2). Despite large advances
in diagnosis and therapy for the disease, the overall 5-year
survival rate of patients with advanced stage OSCC remains <50%
(3,4);
thus, it is important to investigate effective diagnostic
biomarkers and therapeutic targets for improving the prognosis of
patients with OSCC.
Long non-coding RNAs (lncRNAs), a class of
non-protein coding transcripts >200 nucleotides, have been
identified as vital regulators for OSCC progression (5,6). For
example, the lncRNA colon cancer-associated transcript 1 (CCAT1) is
overexpressed in OSCCs and a higher CCAT1 expression in OSCCs has
been reported to predict poor prognosis (7). Additionally, the lncRNA urothelial
cancer-associated 1 (UCA1) contributes to the progression of OSCC
by regulating the WNT/β-catenin signaling pathway (8). Furthermore, the lncRNA
metastasis-associated lung adenocarcinoma transcript 1 promotes
tumor growth and metastasis by inducing epithelial-mesenchymal
transition in OSCC (9). The lncRNA
UCA1 promotes proliferation and cisplatin resistance of OSCC by
suppressing microRNA-184 (miR-184) expression (10). These aforementioned studies indicated
that lncRNAs were involved in OSCC progression.
Small nucleolar RNA host gene 20 (SNHG20), localized
at 17q25.2, has been determined to be involved in a number of tumor
types, including colorectal cancer and gastric cancer. SNHG20
expression is significantly upregulated in colorectal cancer and
increases cell proliferation (11).
Additionally, SNHG20 promotes gastric cancer progression by
inhibiting p21 expression and regulating the glycogen synthase
kinase-3β/β-catenin signaling pathway (12); however, the role of SNHG20 in OSCC
remains unknown.
In the present study, it was determined that SNHG20
was increased in OSCC tissues, compared with in adjacent non-tumor
tissues. Higher SNHG20 expression predicted a poor overall survival
rate of patients with OSCC. Furthermore, knockdown of SNHG20 in
OSCC cells suppressed proliferation ability; thus, the results
demonstrated that SHNG20 may serve as a predictor and potential
target for OSCC treatment.
Materials and methods
Clinical tissue specimens
Human OSCC tissue specimens and matched adjacent
non-tumor tissue specimens were obtained from 40 patients at the
Department of Stomatology, Putuo Hospital, Shanghai University of
Traditional Chinese Medicine (Shanghai, China) between April 2008
and July 2013. Following resection, tissue specimens were
immediately placed in RNAlater® solution (Qiagen GmbH,
Hilden, Germany) and were then stored in liquid nitrogen for
further analysis. The clinicopathological factors of patients are
summarized in Table I. All patients
were clinically examined and staged according to the
Tumor-Node-Metastasis (TNM) and Union for International Cancer
Control classifications (13). The
follow-up duration is the period from surgery to the last follow-up
date. The protocol of the present study was approved by the Ethics
Committee of Putuo Hospital, Shanghai University of Traditional
Chinese Medicine, and written informed consent was obtained from
all patients.
 | Table I.The association between SNHG20
expression and clinicopathological characteristics in patients with
oral squamous cell carcinoma. |
Table I.
The association between SNHG20
expression and clinicopathological characteristics in patients with
oral squamous cell carcinoma.
|
|
| SNHG20
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristics | Total | Reduced (n=20) | Increased (n=20) | P-value |
|---|
| Age |
|
|
| 0.525 |
| ≤60 | 22 | 12 | 10 |
|
|
>60 | 18 | 8 | 10 |
|
| Sex |
|
|
| 0.519 |
|
Female | 16 | 7 | 9 |
|
| Male | 24 | 13 | 11 |
|
| Smoking status |
|
|
| 0.527 |
| No | 20 | 11 | 9 |
|
| Yes | 20 | 9 | 11 |
|
| Tumor site |
|
|
| 0.744 |
|
Tongue | 15 | 8 | 7 |
|
|
Non-tongue | 25 | 12 | 13 |
|
| T stage |
|
|
| 0.197 |
|
T1-T2 | 20 | 14 | 6 |
|
|
T3-T4 | 20 | 6 | 14 |
|
| Differentiation
(13) |
|
|
| 0.025a |
| Well and
moderately | 23 | 15 | 8 |
|
|
Poorly | 17 | 5 | 12 |
|
| Lymph node
metastasis |
|
|
| 0.185 |
| No | 26 | 15 | 11 |
|
| Yes | 14 | 5 | 9 |
|
| TNM stage (13) |
|
|
| 0.011a |
| I–II | 22 | 15 | 7 |
|
|
III–IV | 18 | 5 | 13 |
|
Cell lines culture
A total of three human OSCC cell lines (SCC4, HN4
and SCC15) and the normal human oral keratinocyte (NHOK) cell line
were purchased from the Institute of Biochemistry and Cell Biology
of the Chinese Academy of Sciences (Shanghai, China). Cells were
maintained in Dulbecco's modified Eagle medium supplemented with
10% fetal bovine serum (both from HyClone; GE Healthcare Life
Sciences, Logan, UT, USA). All cells were cultured at 37°C in a
humidified air atmosphere containing 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues and cell lines,
including HN4SCC4, HN4, SCC15 and NHOK cells with the
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). The cDNA was produced by reverse
transcribing 100 ng total RNA. Reverse transcription assay was
conducted to obtain the complementary DNA (cDNA) by Prime Script RT
reagent kit (Takara Biotechnology Co., Ltd., Dalian, China),
according to the manufacturer's protocols. The relative expression
of SNHG20 was detected with the SYBR® Green Master Mix
(Takara Bio, Inc., Otsu, Japan) using a Step One Plus Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). GAPDH
was used as the endogenous control. The relative expression
quantification of SNHG20 was calculated using 2−ΔΔCq
method (14). The thermocycling
conditions for qPCR were as follows: 95°C for 3 min, followed by 40
cycles of 95°C for 12 sec and 58°C for 40 sec. The primers were
synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The
primer sequences were as follows: SNHG20, forward,
5′-ATGGCTATAAATAGATACACGC-3′, and reverse,
5′-GGTACAAACAGGGAGGGA-3′; and GAPDH, forward,
5′-ACAGTCAGCCGCATCTTCT-3′ and reverse,
5′-GACAAGCTTCCCGTTCTCAG-3′.
Cells transfection
A total of two small interfering RNAs (siRNAs)
against SNHG20 were purchased from Shanghai GenePharma Co., Ltd.
The siRNAs sequences were as follows: si-NC,
5′-GGATACGGAGTACTATAGC-3′; si-SNHG20-1,
5′-GCCUAGGAUCAUCCAGGUUTT-3′; and si-SNHG20-2,
5′-GCCACUCACAAGAGUGUAUTT-3′. When cell confluence reached 80–90%,
100 nm si-negative control (si-NC), si-SNHG20-1 or si-SNHG20-2 were
transfected into SCC4 or SCC15 cells using the
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Cells
were harvested at 48 h after transfection.
Cell proliferation assay
Cell proliferation capacity was detected with a Cell
Counting kit-8 (CCK-8; Beyotime Institute of Biotechnology,
Nantong, China). Subsequently, SCC4 and SCC15 cells were seeded
into 96-well plates (2×103 cells/well), and transfected
with si-NC, si-SNHG20-1 or si-SNHG20-2. At 1, 2, 3 and 4 days after
cell transfection, cells were stained using 10 µl CCK-8 reagent.
After 2 h of incubation at 37°C, cell proliferation capacity was
measured using a microplate reader (Enspire 2300 Maltilabel Reader;
PerkinElmer, Inc., Waltham, MA, USA), and the absorbance of samples
was measured at 450 nm.
Colony formation assay
The transfected cells from each group were seeded
into 12-well plates (5×102 cells/well) and cultured for
14 days at 37°C in a humidified air atmosphere containing 5%
CO2. Subsequently, the colonies were then fixed with
100% methanol at room temperature for 20 min and stained with 0.1%
crystal violet at room temperature for 20 min. Finally, cells were
counted and imaged under a X71 fluorescence microscope
(magnification, ×200; Olympus Corporation, Tokyo, Japan).
Western blot analysis
SCC4 and SCC15 cells were lysed with
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology) and quantified using a bicinchoninic acid protein
assay kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) according
to the manufacturer's protocols. A. total of 20 µg total protein
lysates were separated on 10% SDS-PAGE electrophoresis and then
were transferred to polyvinylidene fluoride membranes (EMD
Millipore, Billerica, MA, USA). The membranes were blocked using 5%
free-fat milk for 1 h at room temperature and then were incubated
with anti-proliferating cell nuclear antigen (PCNA) (cat. no.
ab92552; dilution, 1:3,000; Abcam, Cambridge, UK), anti-Ki67 (cat.
no. ab15580; dilution, 1:3,000; Abcam) and anti-GAPDH (cat. no.
sc-20358, dilution, 1:1,000; Santa Cruz Biotechnology, Inc.,
Dallas, CA, USA) antibodies at 4°C overnight. Subsequently,
membranes were incubated with secondary antibody anti-mouse IgG
conjugated with horseradish peroxidase (dilution, 1:5,000; cat. no.
ab97040; Abcam) for 2 h at room temperature and the protein blots
were visualized with the enhanced chemiluminescent detection system
(GE, Fairfield, CT, USA). Protein bands were visualized using
ImageJ 1.45S software (National Institutes of Health, Bethesda, MD,
USA).
Statistical analysis
Data analysis in the present study was performed
using SPSS 20.0 (IBM Corp., Armonk, NY, USA). Results were
presented as the mean ± standard deviation. The associations
between SNHG20 expression and clinicopathological factors were
analyzed using the χ2 test. The significance of
differences between groups was analyzed with the Student's t-test
or one-way ANOVA followed by a Student-Newman-Keuls-q test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Increased SNHG20 expression is
determined in OSCC tissues and associates with histological
differentiation and TNM stage
The expression of SNHG20 in 40 human OSCC tissue
specimens and matched adjacent non-tumor tissue specimens was
examined. As depicted in Fig. 1A, the
SNHG20 expression was significantly increased in OSCC tissue
specimens, compared with in the matched adjacent non-tumor tissue
specimens (P<0.05). Furthermore, SNHG20 expression was
significantly increased in three OSCC cell lines (SCC4, HN4 and
SCC15), compared with in the NHOK cell line (P<0.05; Fig. 1B). In order to understand the clinical
significance of SNHG20 expression in patients, the association
between SNHG20 expression and clinicopathological factors were
analyzed with the χ2 test. The results demonstrated that
SNHG20 overexpression was significantly associated with a reduced
histological differentiation and advanced TNM stage (III–IV stage;
P<0.05; Table I) (13).
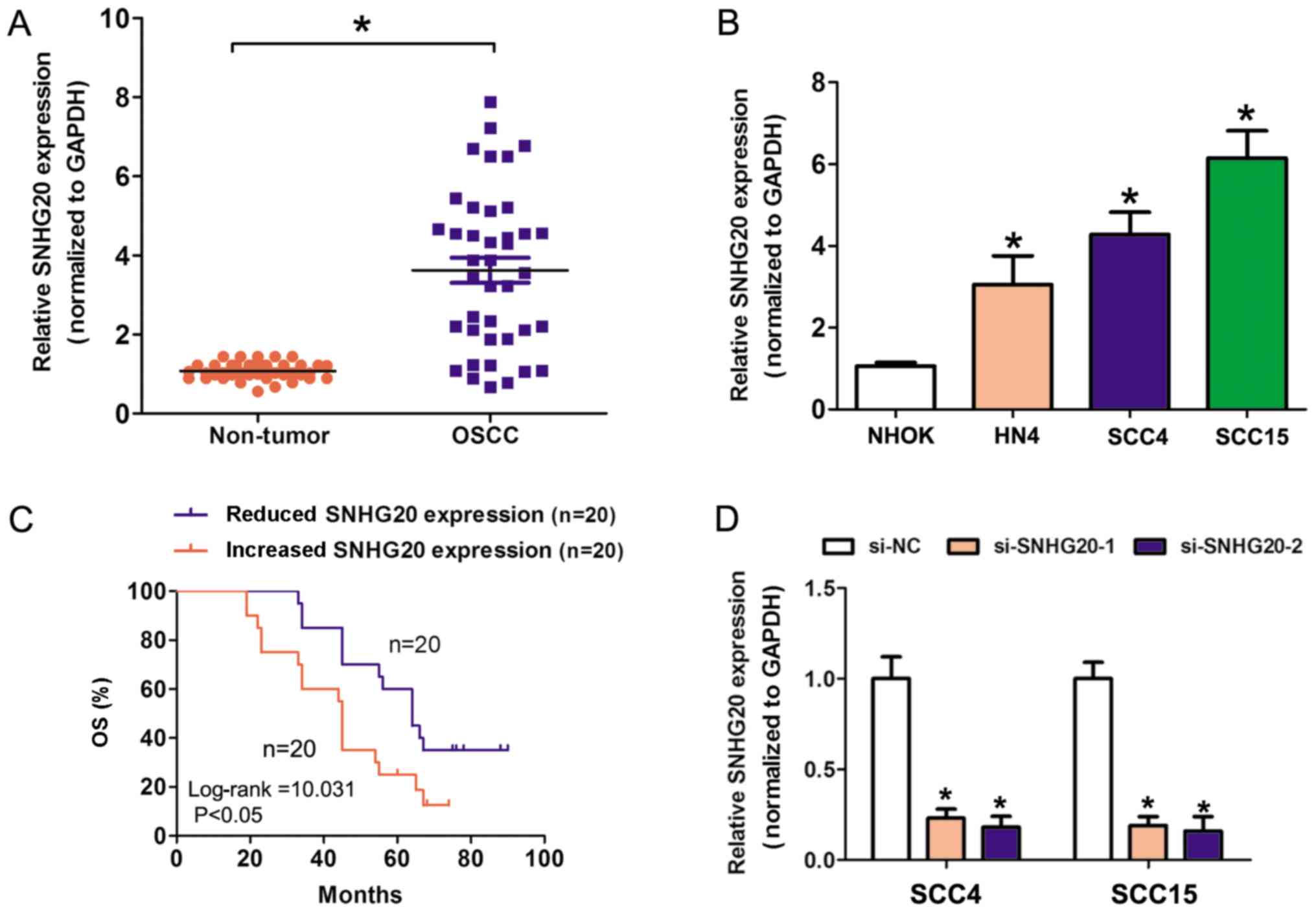 | Figure 1.Increased SNHG20 expression was
determined in OSCC tissues and is associated with a poor OS. (A)
SNHG20 expression was significantly increased in OSCC tissues,
compared with in adjacent non-tumor tissues with a RT-qPCR assay.
(B) SNHG20 expression was significantly increased in three human
OSCC cell lines (SCC4, HN4 and SCC15), compared with in the NHOK
cell line. (C) Kaplan-Meier analysis for the effects of SNHG20
expression on the OS of patients. (D) The relative expression of
SNHG20 was detected with a RT-qPCR assay when SCC4 and SCC15 cells
were transfected with si-NC, si-SNHG20-1 or si-SNHG20-2. Error bars
represent mean ± standard deviation for ≥3 independent experiments,
*P<0.05 compared with si-NC. RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; OSCC, oral
squamous cell carcinoma; SNHG20, small nucleolar RNA host gene 20;
NHOK, normal human oral keratinocyte; si-NC, small
interfering-negative control; OS, overall survival. |
Increased SNHG20 expression is a
predictor of overall survival (OS) of patients of OSCC
Survival plots were performed with the Kaplan-Meier
method and compared with log-rank tests. The results demonstrated
that increased SNHG20 expression in patients with OSCC reduced
their OS, compared with in patients with reduced SNHG20 expression
(Fig. 1C). In univariate analysis,
the results indicated that poor tumor differentiation, advanced
clinical stage and increased SNHG20 expression were independent
predictors of the OS of patients with OSCC, and these associations
were significant (P<0.05; Table
II). Furthermore, multivariate Cox proportional hazards
regression analysis demonstrated that poor tumor differentiation,
advanced clinical stage and increased SNHG20 expression were
independent predictors of the OS of patients with OSCC, and these
associations were significant (P<0.05; Table II); thus, these results indicated
that increased SNHG20 expression may serve as an independent
predictor for the OS of patients with OSCC.
 | Table II.Univariate and multivariate Cox
proportional hazards analysis of SNHG20 expression and OS in
patients with oral squamous cell carcinoma. |
Table II.
Univariate and multivariate Cox
proportional hazards analysis of SNHG20 expression and OS in
patients with oral squamous cell carcinoma.
|
| Univariate Cox
analysis | Multivariate Cox
analysis |
|---|
|
|
|
|
|---|
| Clinicopathological
characteristics | RR (95% CI) | P-value | RR (95% CI) | P-value |
|---|
| Age | 0.788
(0.355–1.477) | 0.655 |
|
|
| Sex | 1.088
(0.546–1.877) | 0.565 |
|
|
| Smoking status | 0.927
(0.443–1.788) | 0.588 |
|
|
| Tumor site | 1.156
(0.577–1.993) | 0.422 |
|
|
| Non-tongue | 0.655
(0.301–1.444) | 0.723 |
|
|
| T stage | 0.756
(0.404–1.466) | 0.688 |
|
|
| Differentiation
(13) | 2.131
(1.587–2.977) | 0.001a | 1.896
(1.448–2.745) | 0.002a |
| Lymph node
metastasis | 1.223
(0.779–1.987) | 0.332 |
|
|
| TNM stage (13) | 2.289
(1.642–3.224) | 0.001a | 1.966
(1.246–3.009) | 0.001a |
| SNHG20 | 2.556
(1.822–3.663) | 0.001a | 2.077
(1.388–3.244) | 0.001a |
Decreased expression of SNHG20
expression inhibits cell proliferation
The impact of SNHG20 expression on OSCC cell
proliferation was also examined. SNHG20 expression was knocked down
in two increased SNHG20 expression OSCC cell lines (SCC4 and SCC15
cells). The knockdown efficiency of two siRNAs was increased and
depicted in Fig. 1D. The cell
proliferation ability was detected using CCK-8 cell proliferation
and cell colony formation assays. The CCK-8 results indicated that
decreased expression of SNHG20 significantly inhibited the cell
proliferation capacity of SCC4 and SCC15 cells, compared with their
respective negative control groups (P<0.05; Fig. 2A and B). Additionally, the colony
forming assay results demonstrated that decreased expression of
SNHG20 significantly reduced the number of SCC4 and SCC15 cell
colonies, compared with their respective negative control groups
(P<0.05; Fig. 2C and D). It was
also indicated that knockdown of SNHG20 downregulated the
expression of PCNA and Ki67 in SCC4 and SCC15 cells, compared with
their respective negative control groups (Fig. 3A-D). The present results demonstrated
that decreased expression of SNHG20 inhibited cell
proliferation.
Discussion
Recent evidence demonstrated that lncRNAs act as
regulatory molecules that mediate cellular processes, including
chromatin remodeling, transcription, post-transcriptional
modifications and signal transduction (15,16).
lncRNAs could affect cell proliferation, cell apoptosis,
differentiation and invasion, which serves crucial roles in
biological processes (17,18). lncRNAs in OSCC progression could
provide diagnostic and therapeutic strategies for this disease.
SNHG20, a recently determined RNA molecule, is identified as an
oncogene in a number of tumor types including colorectal cancer and
gastric cancer. However, to the best of our knowledge, the
prognostic value and functional role of SHNG20 in OSCC remains
unknown. In the present study, the results indicated that SNHG20
expression was increased in OSCC tissue specimens, compared with in
adjacent non-tumor tissue specimens. SNHG20 overexpression was
associated with reduced histological differentiation and advanced
clinical stage. Multivariate Cox proportional hazards regression
analysis demonstrated that increased SNHG20 expression was an
independent predictor for the OS of patients with OSCC.
In the previous study, SNHG20 was determined to be
increased in colorectal cancer tissues, compared with in
corresponding non-tumor tissues, and high expression of SNHG20 was
notably associated with advanced TNM stage in patients with
colorectal cancer (11).
Additionally, upregulation of SNHG20 predicts a poor overall
survival rate in patients with hepatocellular carcinoma. Knockdown
of SNHG20 in SK-Hep-1 cells significantly inhibited cellular
proliferation, migration and invasion (19). SNHG20 promotes cell proliferation,
invasion and migration of breast cancer cells via regulating
miR-495 (20). Consistently, the
present results demonstrated that decreased expression of SNHG20
expression inhibited cellular proliferation capacity and cell
colony formation ability. Additionally, it was indicated that
decreased expression of SNHG20 downregulated the expression of PCNA
and Ki67 in OSCC cells. PCNA and Ki67 are cell proliferation
markers, whose rate of synthesis is directly associated with the
rates of cellular proliferation and DNA synthesis (21). The present results indicated that
SNHG20 may affect the PCNA and Ki67 expression by potential
mechanisms. These results demonstrated that decreased SNHG20
expression inhibited cell proliferation. Furthermore, in the future
in vivo experiments should be performed, in order to
demonstrate the role of SNHG20 in OSCC.
In conclusion, in the present study, it was
demonstrated that SNHG20 expression was elevated in OSCC tissues
and cells. Increased SNHG20 expression functions as a prognostic
predictor for OSCC tissues. Furthermore, it was demonstrated that
decreased SNHG20 expression inhibited cell proliferation of OSCC;
thus, these results indicated that SNHG20 may act as a prognostic
predictor for OSCC and a target of OSCC treatment. Additionally,
the precise molecular mechanisms underlying OSCC progression
require further investigation.
Acknowledgements
Not applicable.
Funding
No funding received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
PG contributed to the design of the study and was
responsible for project design, organization, manuscript writing,
the supervision of experimental progress and final approval of the
version to be published. PG, RF and TG conducted the molecular
experiments. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The protocol of the present study was approved by
the Ethics Committee of Putuo Hospital, Shanghai University of
Traditional Chinese Medicine, and written informed consent was
obtained from all patients.
Patient consent for publication
Written informed consent was obtained from all
patients and consent for publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Warnakulasuriya S: Global epidemiology of
oral and oropharyngeal cancer. Oral Oncol. 45:309–316. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bourhis J, Overgaard J, Audry H, Ang KK,
Saunders M, Bernier J, Horiot JC, Le Maître A, Pajak TF, Poulsen
MG, et al: Hyperfractionated or accelerated radiotherapy in head
and neck cancer: A meta-analysis. Lancet. 368:843–854. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Leemans CR, Braakhuis BJ and Brakenhoff
RH: The molecular biology of head and neck cancer. Nat Rev Cancer.
11:9–22. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hunter KD, Parkinson EK and Harrison PR:
Profiling early head and neck cancer. Nat Rev Cancer. 5:127–135.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gomes CC, de Sousa SF, Calin GA and Gomez
RS: The emerging role of long noncoding RNAs in oral cancer. Oral
Surg Oral Med Oral Pathol Oral Radiol. 123:235–241. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
González-Ramírez I, Soto-Reyes E,
Sánchez-Pérez Y, Herrera LA and García-Cuellar C: Histones and long
non-coding RNAs: The new insights of epigenetic deregulation
involved in oral cancer. Oral Oncol. 50:691–695. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Arunkumar G, Murugan AK, Prasanna
Srinivasa Rao H, Subbiah S, Rajaraman R and Munirajan AK: Long
non-coding RNA CCAT1 is overexpressed in oral squamous cell
carcinomas and predicts poor prognosis. Biomed Rep. 6:455–462.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang YT, Wang YF, Lai JY, Shen SY, Wang F,
Kong J, Zhang W and Yang HY: Long non-coding RNA UCA1 contributes
to the progression of oral squamous cell carcinoma by regulating
the WNT/β-catenin signaling pathway. Cancer Sci. 107:1581–1589.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren
Y, Wu Y, Mei M, Zhang L and Wang X: Long non coding RNA MALAT1
promotes tumor growth and metastasis by inducing
epithelial-mesenchymal transition in oral squamous cell carcinoma.
Sci Rep. 5:159722015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fang Z, Zhao J, Xie W, Sun Q, Wang H and
Qiao B: LncRNA UCA1 promotes proliferation and cisplatin resistance
of oral squamous cell carcinoma by sunppressing miR-184 expression.
Cancer Med. 6:2897–2908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li C, Zhou L, He J, Fang XQ, Zhu SW and
Xiong MM: Increased long noncoding RNA SNHG20 predicts poor
prognosis in colorectal cancer. BMC Cancer. 16:6552016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu J, Liu L, Wan JX and Song Y: Long
noncoding RNA SNHG20 promotes gastric cancer progression by
inhibiting p21 expression and regulating the GSK-3β/β-catenin
signaling pathway. Oncotarget. 8:80700–80708. 2017.PubMed/NCBI
|
|
13
|
Wahi PN, Cohen B, Luthra UK and Torloni H:
Histological considerationHistological typing of oral and
oropharyngeal tumors. World Health Organization; Geneva,
Switzerland: pp. 15–19. 1971
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Anastasiadou E, Jacob LS and Slack FJ:
Non-coding RNA networks in cancer. Nat Rev Cancer. 18:5–18. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
McMahon M, Samali A and Chevet E:
Regulation of the unfolded protein response by noncoding RNA. Am J
Physiol Cell Physiol. 313:C243–C254. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Meller VH, Joshi SS and Deshpande N:
Modulation of chromatin by noncoding RNA. Annu Rev Genet.
49:673–695. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cheetham SW, Gruhl F, Mattick JS and
Dinger ME: Long noncoding RNAs and the genetics of cancer. Br J
Cancer. 108:2419–2425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang D, Cao C, Liu L and Wu D:
Up-regulation of LncRNA SNHG20 Predicts Poor Prognosis in
Hepatocellular Carcinoma. J Cancer. 7:608–617. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guan YX, Zhang ZM, Chen XZ, Zhang Q, Liu
SZ and Zhang YL: Lnc RNA SNHG20 participated in proliferation,
invasion and migration of breast cancer cells via miR-495. J Cell
Biochem. 119:7971–7981. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Coşarcă AS, Mocan SL, Păcurar M, Fülöp E
and Ormenişan A: The evaluation of Ki67, p53, MCM3 and PCNA
immunoexpressions at the level of the dental follicle of impacted
teeth, dentigerous cysts and keratocystic odontogenic tumors. Rom J
Morphol Embryol. 57:407–412. 2016.PubMed/NCBI
|

















