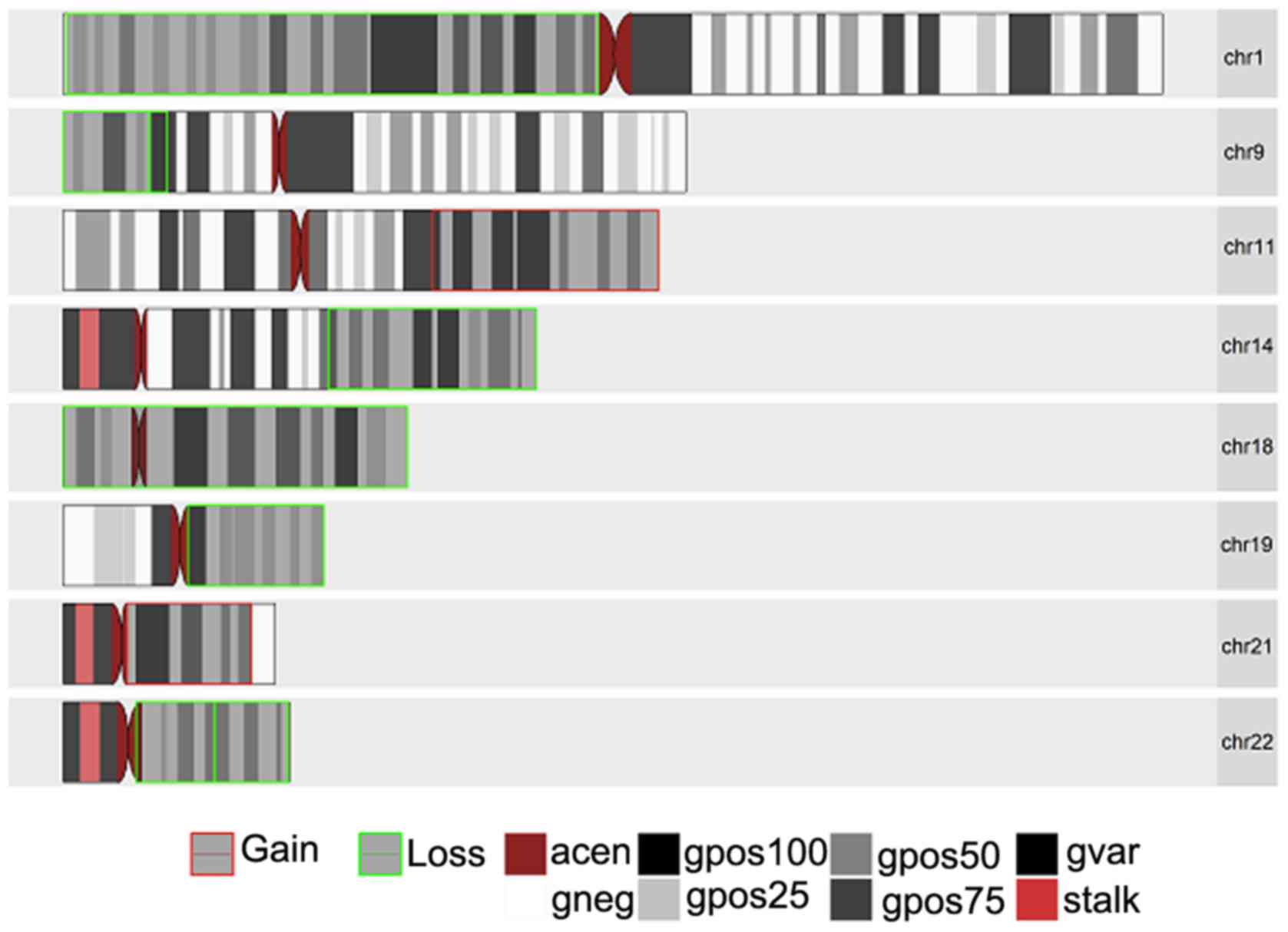
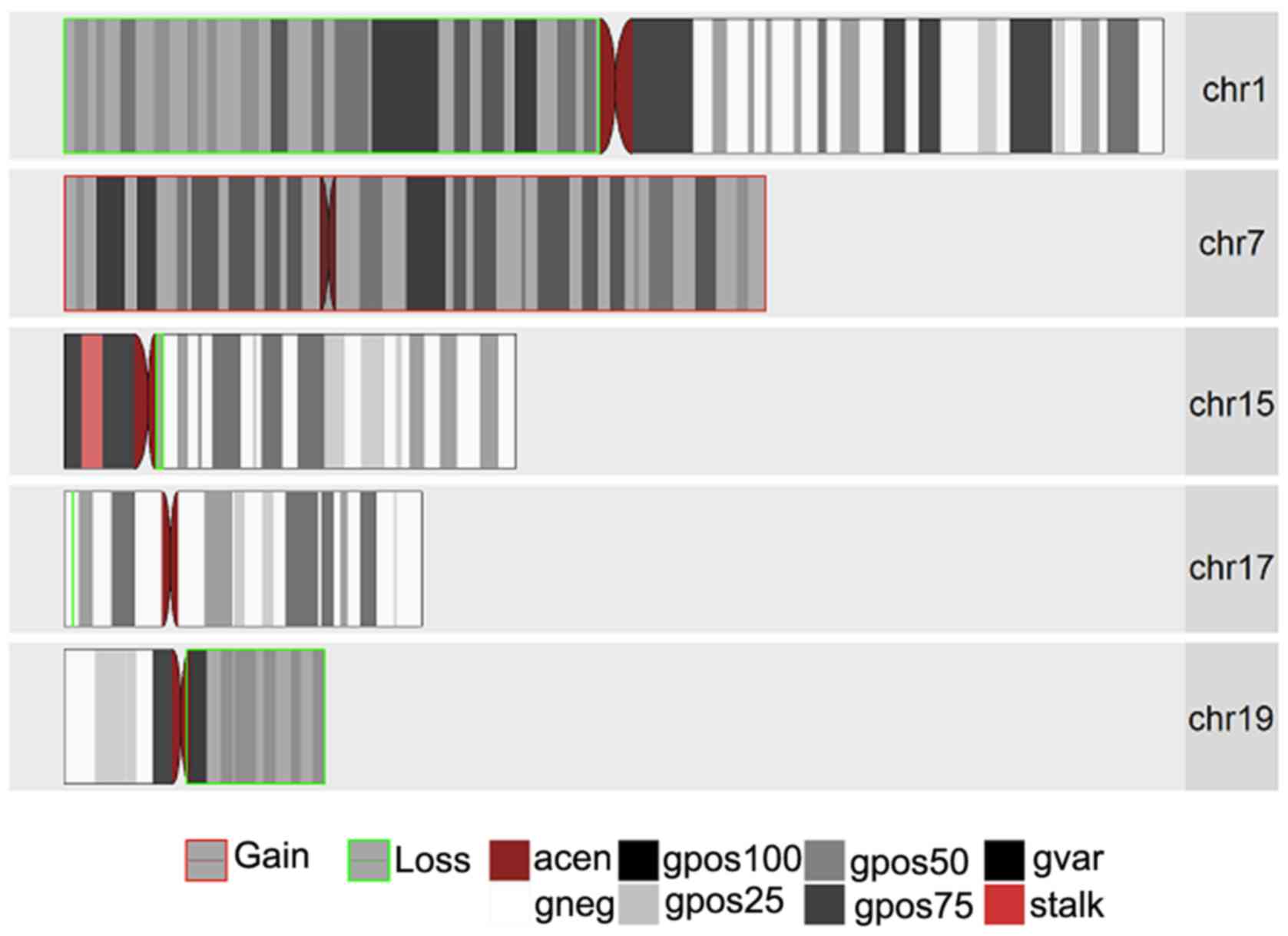
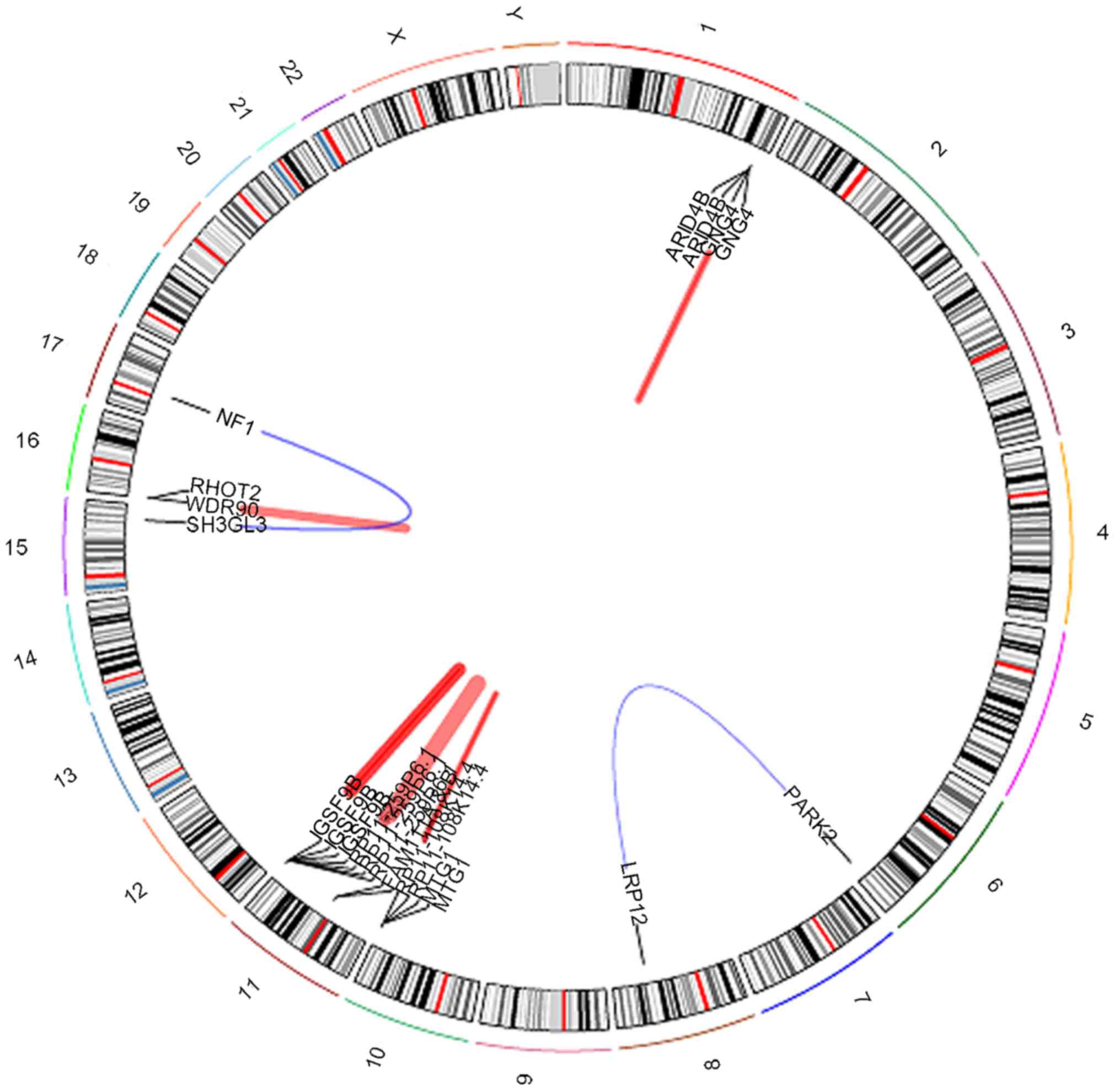
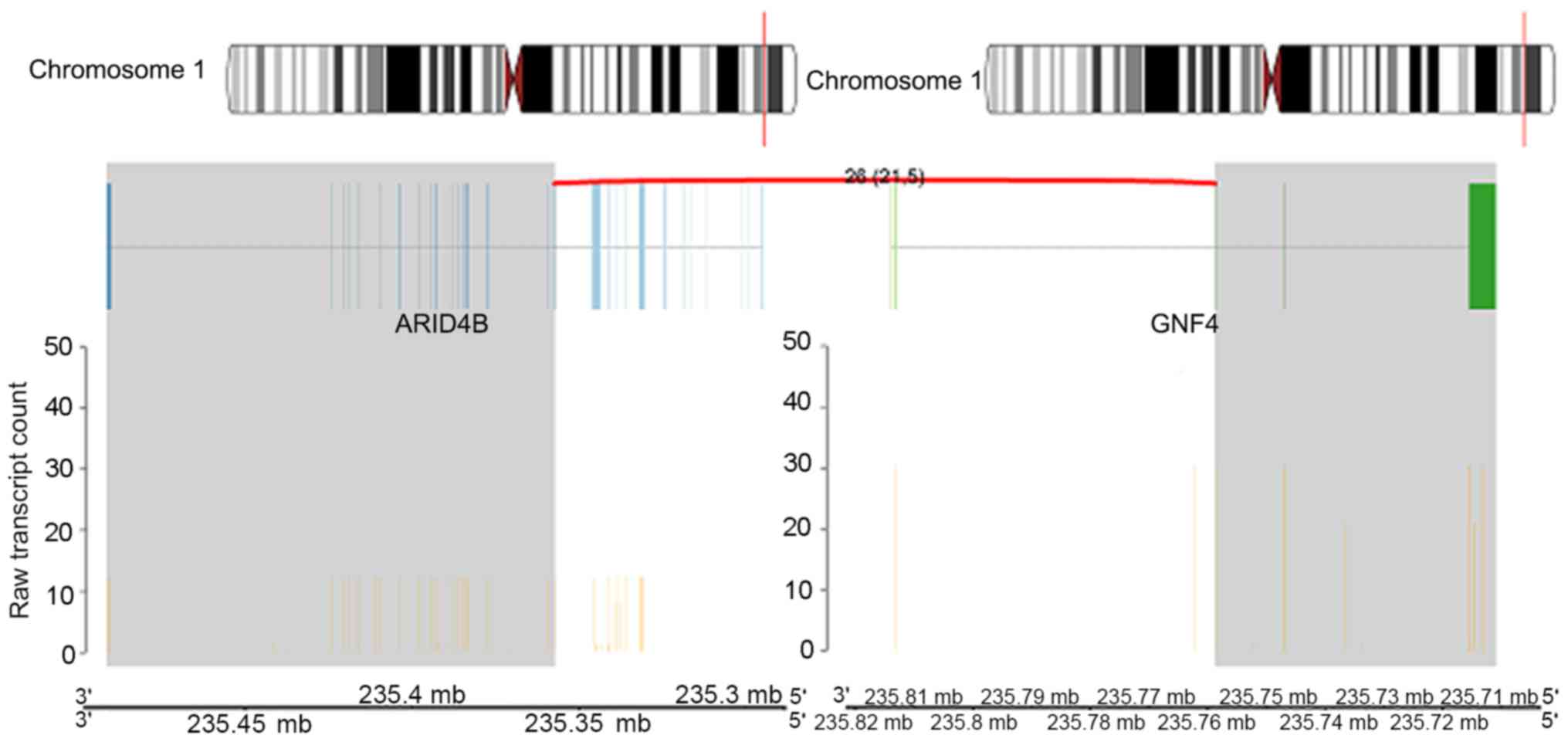
|
1
|
Stewart BW and Wild CP: World cancer
report. 2014, International agency for research on cancer (IARC)
Publications; Lyon, France: 2014, http://www.iarc.fr/en/publications/books/wcr/index.php20–01.
2018
|
|
2
|
Ferlay J, Soerjomataram I, Ervik M,
Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and
Bray F: GLOBOCAN 2012 v1.0, Cancer incidence and mortality
worldwide. IARC CancerBase no.11. IARC Press; Lyon: 2013
|
|
3
|
US Mortality Data: 2006. National Centre
for Health Statistics. Centres for Disease Control and Prevention.
2009.
|
|
4
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihaus P: The 2007
WHO classification of tumors of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang X, Zhang W, Cao WD, Cheng G and
Zhang YQ: Glioblastoma multiforme: Molecular characterization and
current treatment strategy (Reviw). Exp Ther Med. 3:9–14. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li J, Miao N, Liu M, Ciu W, Liu X, Shi X,
Qing S, Ma Y, Zhang W and Biekemituofu H: Clinical significance of
chromosome 1p/19q loss of heterozygosity and Sox17 expression in
oligodendrogliomas. Int J Clin Exp Pathol. 7:8609–8615.
2014.PubMed/NCBI
|
|
7
|
Kakkar A, Suri V, Jha P, Srivasta A,
Sharma V, Pathak P, Sharma MC, Sharma MS, Kale SS, Chosdol K, et
al: Loss of heterozygosity on chromosome 10q in glioblastomas, and
its association with other genetic alterations and survival in
Indian patients. Neurol India. 59:254–261. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hartmann C, Hentschel B, Tatagiba M,
Schramm J, Schnell O, Seidel C, Stein R, Reifenberger G, Pietsch T,
von Deimling A, et al: Molecular markers in low-grade gliomas:
Predictive or prognostic? Clin Cancer Res. 17:4588–4599. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Burger PC, Minn AY, Smith JS, Borell TJ,
Jedlicka AE, Huntley BK, Goldthwaite PT, Jenkins RB and Feuerstein
BG: Losses of chromosomal arms 1p and 19q in the diagnosis of
oligodendroglioma. A study of paraffin-embedded sections. Mod
Pathol. 14:842–853. 2001.
|
|
10
|
Haynes HR, Camelo-Piragua S and Kurian KM:
Prognostic and predictive biomarkers in adult and pediatric
gliomas: Toward personalized treatment. Front Oncol. 4:472014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gadji M, Fortin D, Tsanaclis AN and Drouin
R: Is the 1p/19q deletion a diagnostic marker of
oligodendrogliomas? Cancer Genet Cytogenet. 194:12–22. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hartmann C, Meyer J, Balss J, Capper D,
Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, et
al: Type and frequency of IDH1 and IDH2 mutations are related to
astrocytic and oligodendroglial differentiation and age: A study of
1,010 diffuse gliomas. Acta Neuropathol. 118:469–474. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nayak A, Ralte AM, Sharma MC, Singh VP,
Mahapatra AK, Mehta VS and Sarkar C: p53 protein alterations in
adult astrocytic tumors and oligodendrgliomas. Neurol India.
52:228–232. 2004.PubMed/NCBI
|
|
14
|
Blakely J and Grossman S: Anaplastic
oligodendroglioma. Curr Treat Options Neurol. 10:295–307. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mohapatra G, Betensky RA, Miller ER, Carey
B, Gaumont LD, Engler DA and Loius DN: Glioma test array for use
with formalin-fixed, paraffin-embedded tissue: Array comparative
genomic hybridization correlates with loss of heterozygosity and
fluorescence in situ hybridization. J Mol Diagn. 8:268–276. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Meyerson M, Gabriel S and Getz G: Advances
in understanding cancer genomes through second-generation
sequencing. Nat Rev Genet. 11:685–696. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Robinson K: Application of
second-generation sequencing to cancer genomics. Brief Bioinform.
11:524–534. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Patil V, Pal J and Somasundaram K:
Elucidating the cancer-specific genetic alteration spectrum of
glioblastoma derived cell lines from whole exome and RNA
sequencing. Oncotarget. 6:43452–43471. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Chen K, Sloan SA, Bennet ML,
Scholze AR, O'Keefe S, Phatnani HP, Guarnieri P, Caneda C,
Ruderisch N, et al: An RNA-sequencing transcriptome and splicing
database of glia, neurons, and vascular cells of the cerebral
cortex. J Neurosci. 34:11929–11947. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao Z, Meng F, Wang W, Wang Z, Zhang C
and Jiang T: Comprehensive RNA-seq transcriptomic profiling in the
malignant progression of gliomas. Sci Data. 4:1700242017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Andrews S: FastQC: A quality control tool
for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc15–06.
2017
|
|
22
|
Flicek P, Ahmed I, Amode MR, Barrell D,
Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S,
et al: Ensembl 2013. Nucleic Acids Res. 41:(Database Issue).
D48–D55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Trapnell C, Pachter L and Salzberg SL:
Tophat: Discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzburg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Trapnell C, Williams BA, Pertea G,
Mortazavi A, Kwan G, van Baren MJ, Salzburg SL, Wold BJ and Pachter
L: Transcript assembly and quantification by RNA-Seq reveals
unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bello MJ, Vaquero J, de Campos JM, Kusak
ME, Sarasa JL, Saez-Castresana J, Pestana A and Rey JA: Molecular
analysis of chromosome 1 abnormalities in human gliomas reveals
frequent loss of 1p in oligodendroglial tumors. Int J Cancer.
57:172–175. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bello MJ, Leone PE, Vaquero J, de Campos
JM, Kusak ME, Sarasa JL, Pestaña A and Rey JA: Allelic loss at 1p
and 19q frequently occurs in association and may represent early
oncogenic events in oligodendroglial tumors. Int J Cancer.
64:207–210. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cairncross JG, Ueki K, Zlatescu MC, Lisle
DK, Finkelstein DM, Hammond RR, Silver JS, Stark PC, Macdonald DR,
Ino Y, et al: Specific genetic predictors of chemotherapeutic
response and survival in patients with anaplastic
oligodendrogliomas. J Natl Cancer Inst. 90:1473–1479. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baumann GS, Ino Y, Ueki K, Zlatescu MC,
Fisher BJ, Macdonald DR, Stitt L, Louis DN and Cairncross JG:
Allelic loss of chromosome 1p and radiotherapy plus chemotherapy in
patients with oligodendrogliomas. Int J Radiat Oncol Biol Phys.
48:825–830. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hagel C, Krog B, Laas R and Stavrou DK:
Prognostic relevance of TP53 mutations, p53 protein, Ki-67 index
and conventional histologic grading in oligodendrogliomas. J Exp
Clin Cancer Res. 18:305–309. 1999.PubMed/NCBI
|
|
31
|
Korshunov A and Golanov A: The prognostic
significance of vascular endothelial growth factor (VEGF C-1)
immunoexpression in oligodendroglioma. An analysis of 91 cases. J
Neurooncol. 48:13–19. 2000. View Article : Google Scholar
|
|
32
|
Di Rocco F, Carroll RS, Zhang J and Black
PM: Platelet-derived growth factor and its receptor expression in
human oligodendrogliomas. Neurosurg. 42:341–346. 1998. View Article : Google Scholar
|
|
33
|
Iwabuchi H, Sakamoto M, Sakunaga H, Ma YY,
Carcangiu ML, Pinkel D, Yang-Feng TL and Gray JW: Genetic analysis
of benign, low-grade, and high-grade ovarian tumors. Cancer Res.
55:6172–6180. 1995.PubMed/NCBI
|
|
34
|
Parker BC, Annala MJ, Cogdell DE, Granberg
KJ, Sun Y, Ji P, Li X, Gumin J, Zheng H, Hu L, et al: The
tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in
glioblastoma. J Clin Invest. 123:855–865. 2013.PubMed/NCBI
|
|
35
|
Singh D, Chan JM, Zoppoli P, Niola F,
Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S,
et al: Transforming fusions of FGFR and TACC genes in human
glioblastoma. Science. 337:1231–1235. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jones DT, Kocialkowski S, Liu L, Pearson
DM, Bäcklund LM, Ichimura K and Collins VP: Tendem duplication
producing a novel oncogenic BRAF fusion gene defines the majority
of pilocytic astrocytomas. Cancer Res. 68:8673–8677. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cao J, Gao T, Stanbridge EJ and Irie R:
RBP1L1, a retinoblastomabinding protein-related gene encoding an
antigenic epitope abundantly expressed in human carcinomas and
normal testis. J Natl Cancer Inst. 93:1159–1165. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Neidhart M: Chapter 14-DNA Methylation in
Growth Retardation. In DNA Methylation and Complex Human Disease.
Academic Press; Oxford: pp. 241–259. 2016, View Article : Google Scholar
|
|
39
|
Goldberger N, Walker RC, Kim CH, Winter S
and Hunter KW: Inherited variation in miR-290 expression suppresses
breast cancer progression by targeting the metastasis
susceptibility gene Arid4b. Cancer Res. 73:2671–2681. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu MY, Eldin KW and Beaudet AL:
Identification of chromatin remodeling genes Arid4a and Arid4b as
leukemia suppressor genes. J Natl Cancer Inst. 100:1247–1259. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tsai WC, Hueng DY, Nieh S and Gao HW:
ARID4B is a good biomarker to predict tumour behaviour and decide
WHO grades in gliomas and meningiomas. J Clin Pathol. 70:162–167.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pal J, Patil V, Mondal B, Shukla S, Hegde
AS, Arivazhagan A, Santosh V and Somasundaram K: Epigenetically
silenced GNG4 inhibits SDF1α/CXCR4 signaling in mesenchymal
glioblastoma. Genes Cancer. 7:136–147. 2016.PubMed/NCBI
|
|
43
|
Mercier S, Küry S, Shaboodien G, Hounier
DT, Khumalo NP, Bou-Hanna C, Bodak N, Cormier-Daire V, David A,
Faivre L, et al: Mutations in FAM111B cause hereditary fibrosing
poikiloderma with tendon contracture, myopathy, and pulmonary
fibrosis. Am J Hum Genet. 93:1100–1107. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Goussot R, Prasad M, Stoetzel C, Lenormand
C, Dollfus H and Lipsker D: Expanding phenotype of hereditary
fibrosing poikiloderma with tendon contractures, myopathy, and
pulmonary fibrosis caused by FAM111B mutations: Report of an
additional family raising the question of cancer predisposition and
a short review of early-onset poikiloderma. JAAD Case Rep.
3:143–150. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Unger S, Górna MW, Le Béchec A, Do
Vale-Pereira S, Bedeschi MF, Geiberger S, Grigelioniene G,
Horemuzova E, Lalatta F, Lausch E, et al: FAM111A mutations result
in hypoparathyroidism and impaired skeletal development. Am J Hum
Genet. 92:990–995. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Stirnimann CU, Petsalaki E, Russell RB and
Müller CW: WD40 proteins propel cellular networks. Trends Biochem
Sci. 35:565–574. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Fransson A, Ruusala A and Aspenström P:
Atypical Rho GTPases have roles in mitochondrial homeostasis and
apoptosis. J Biol Chem. 278:6495–6502. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fransson S, Ruusala A and Aspenström P:
The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in
mitochondrial trafficking. Biochem Biophys Res Commun. 344:500–510.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Caino MC, Seo JH, Aguinaldo A, Wai E,
Bryant KG, Kossenkov AV, Hayden JE, Vaira V, Morotti A, Ferrero S,
et al: A neuronal network of mitochondrial dynamics regulates
metastasis. Nat Commun. 7:137302016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Garnis C, Coe BP, Zhang L, Rosin MP and
Lam WL: Overexpression of LRP12, a gene contained within an 8q22
amplicon identified by high-resolution array CGH analysis of oral
squamous cell carcinomas. Oncogene. 23:2582–2586. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bethge N, Honne H, Andresen K, Hilden V,
Trøen G, Liestøl K, Holte H, Delabie J, Lind GE and Smeland EB: A
gene panel, including LRP12, is frequently hypermethylated in major
types of B-cell lymphoma. PLoS One. 9:e1042492014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Robens BK, Gembé E, Fassunke J, Becker AJ,
Schoch S and Grote A: Abundance of LRP12 C-rs9694676 allelic
promoter variant in epilepsy-associated gangliogliomas. Life Sci.
155:70–75. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gong Y, Schumacher SE, Wu WH, Tang F,
Beroukhim R and Chan TA: Pan-cancer analysis links PARK2 to
BCL-XL-dependent control of apoptosis. Neoplasia. 19:75–83. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Andersen LB, Ballester R, Marchuk DA,
Chang E, Gutmann DH, Saulino AM, Camonis J, Wigle M and Collins FS:
A conserved alternative splice in the von Recklinghausen
neurofibromatosis (NF1) gene produces two neurofibromin isoforms,
both of which have GTPase-activating protein activity. Mol Cell
Biol. 13:487–495. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Banerjee S, Crouse NR, Emnett RJ, Gianino
SM and Gutmann DH: Neurofibromatosis-1 regulates mTOR-mediated
astrocyte growth and glioma formation in a TSC/Rheb-independent
manner. Proc Natl Acad Sci USA. 108:15996–16001. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Szudek J, Birch P, Riccardi VM, Evans DG
and Friedman JM: Associations of clinical features in
neurofibromatosis 1 (NF1). Genet Epidemiol. 19:429–439. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gutmann DH, Rasmussen SA, Wolkenstein P,
MacCollin MM, Guha A, Inskip PD, North KN, Poyhonen M, Birch PH and
Friedman JM: Gliomas presenting after age 10 in individuals with
neurofibromatosis type 1 (NF1). Neurology. 59:759–761. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Nielsen GP, Stemmer-Rachamimov AO, Ino Y,
Moller MB, Rosenberg AE and Louis DN: Malignant transformation of
neurofibromas in neurofibromatosis 1 is associated with CDKN2A/p16
inactivation. Am J Pathol. 155:1879–1884. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Legius E, Dierick H, Wu R, Hall BK,
Marynen P, Cassiman JJ and Glover TW: TP53 mutations are frequent
in malignant NF1 tumors. Genes Chromosomes Cancer. 10:250–255.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Vizcaíno MA, Shah S, Eberhart CG and
Rodriguez FJ: Clinico-pathologic implications of NF1 gene
alterations in diffuse gliomas. Human Pathology. 46:1323–1330.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bruzek AK, Zureick AH, McKeever PE, Garton
HJL, Robertson PL, Mody R and Koschmann CJ: Molecular
characterization reveals NF1 deletions and FGFR1-activating
mutations in a pediatric spinal oligodendroglioma. Pediatr Blood
Cancer. 64:e263462017. View Article : Google Scholar
|
|
62
|
Giachino C, Lantelme E, Lanzetti L,
Saccone S, Bella Valle G and Migone N: A Novel SH3-containing human
gene family preferentially expressed in the central nervous system.
Genomics. 41:427–434. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sittler A, Walter S, Wedemeyer N,
Hasenbank R, Scherzinger E, Eickhoff H, Bates GP, Lehrach H and
Wanker EE: SH3GL3 associates with the huntingtin exon 1 protein and
promotes the formation of polygln-containing protein aggregates.
Mol Cell. 2:427–436. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Huebner K, Kastury K, Druck T, Salcini AE,
Lanfrancone L, Pelicci G, Lowenstein E, Li W, Park SH, Cannizzaro
L, et al: Chromosome locations of genes encoding human signal
transduction adapter proteins, Nck (NCK), Shc (SHC1), and Grb2
(GRB2). Genomics. 22:281–287. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Fang WJ, Zheng Y, Wu LM, Ke QH, Shen H,
Yuan Y and Zheng SS: Genome-wide analysis of aberrant DNA
methylation for identification of potential biomarkers in
colorectal cancer patients. Asian Pac J Cancer Prev. 13:1917–1921.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chen R, Zhao H, Wu D, Zhao C, Zhao W and
Zhou X: The role of SH3GL3 in myeloma cell migration/invasion,
stemness and chemo-resistance. Oncotarget. 7:73101–73113.
2016.PubMed/NCBI
|
|
67
|
Delic S, Lottmann N, Jetschke K,
Reifenberger G and Riemenschneider MJ: Identification and
functional validation of CDH11, PCSK6 and SH3GL3 as novel glioma
invasion-associated candidate genes. Neuropathol Appl Neurobiol.
38:201–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Aphrothiti JH, Gopa I and David B: Solit:
2-Intracellular Signaling, In Abeloff's Clinical Oncology (Fifth
edition). John EN, James OA, James HD, Michael BK and Joel ET:
Tepper, Content Repository Only; Philadelphia: pp. 22–39. 2014
|
|
69
|
Zhu Y, Zhang X, Wang L, Ji Z, Xie M, Zhou
X, Liu Z, Shi H and Yu R: Loss of SH3GL2 promotes the migration and
invasion behaviours of glioblastoma cells through activating the
STAT3/MMP2 signalling. J Cell Mol Med. 21:2685–2694. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wiederhold T, Lee MF, James M, Neujahr R,
Smith N, Murthy A, Hartwig J, Gusella JF and Ramesh V: Magicin, a
novel cytoskeletal protein associates with the NF2 tumor suppressor
merlin and Grb2. Oncogene. 23:8815–8825. 2004. View Article : Google Scholar : PubMed/NCBI
|