Introduction
Renal cell carcinoma (RCC) is the most common renal
tumor accounting for 2–3% of all malignancies worldwide (1). Several histological RCC subtypes have
been categorized, and the most frequent subtypes include clear cell
RCC (ccRCC), papillary RCC and chromophobe RCC. These subtypes
together represent >90% of all diagnosed RCCs (2). Nephrectomy was historically the only
treatment option for RCC, with no requirement for advanced
prognostication or follow-up. However, since the turn of the
century, there have been dramatic changes in the available
diagnostics and treatment options (3). There is an increasing incidence of
detection of small renal tumors (<40 mm in diameter), primarily
due to the increasing use of tomographic radiology, which enables
for the improved detection of disease (4,5). Small
tumors often grow slowly; however, not all small tumors are
indolent in nature (6). Several
prognostic factors for RCC are presently used in a clinical
setting, including tumor stage, Fuhrman grade, lymph node
involvement and histological subtype. However, these factors lack
accuracy in predicting the natural history of the disease,
particularly in patients with non-metastatic disease at the time of
diagnosis (7).
Overall, 20–30% of all patients with RCC present
with metastatic disease at the time of diagnosis, and 20–40%
develop metastases following nephrectomy (8). The need for prognostic tools in
metastatic (m)RCC has also become evident. There are now several
available targeted therapies on offer to patients with mRCC in
conjunction with metastatic surgery and other ablative therapies of
the metastatic lesions. The prognosis of RCC can vary widely, and
early detection of recurrence can improve patient outcome, as
systemic treatments are more likely to yield a favorable response
when the metastatic burden is limited (9). Therefore, there is a requirement to
identify molecular biomarkers that can aid in predicting patient
outcome for patients with RCC, either alone or in combination with
the presently used clinical parameters.
MicroRNAs (miRNAs/miRs) are small non-coding RNA
molecules that regulate gene expression at a post-transcriptional
level (10). These molecules serve a
key role in a diverse range of biological processes important in
cancer development, including proliferation, differentiation and
apoptosis (11,12). A single miRNA could alter the
expression of a large number of target genes, and therefore has the
potential to regulate entire disease-specific pathways and
signaling cascades (13). Altered
miRNA expression has been identified in all human tumors
investigated to date, and several diagnostic kits based on miRNA
expression profiles are available, including one for the
differentiation between histological subtypes of RCC (available
from Rosetta Genomics, Ltd., Princeton, NJ, USA). Three of the
miRNAs that have previously been suggested as prognostic biomarkers
for RCC are miR-21, miR-126 and miR-10b (14–21).
miR-21 is an oncogenic miRNA that has been identified to be
upregulated in numerous cancer types, including RCC (14,15). The
overexpression of miR-21 increases cell proliferation, migration
and invasion, as well as inhibiting apoptosis, and the confirmed
target genes for miR-21 are tumor protein p53, phosphatase and
tensin homolog, and programmed cell death 4 (22–26). The
deregulation of miR-126 has also been identified in various types
of cancer, where it regulates genes involved in the vascular
endothelial growth factor and phosphatidylinositol 3-kinase
pathways, and therefore serves an important role in processes
including angiogenesis and cell cycle regulation (27–34). The
third miRNA, miR-10b, has previously been reported as an oncomiR in
specific types of cancer, while it has been reported to exert a
tumor suppressor role in others, among them RCC (16,35–38). The
overexpression of miR-10b in RCC cell lines inhibits cell
proliferation, migration and invasion; however, the exact mechanism
has not been completely elucidated (17,38).
Even though the expression of these 3 miRNAs have previously been
investigated in RCC, their use as prognostic biomarkers for this
disease remains debatable. Therefore, further studies are required
in order to validate their potential to predict clinical outcome in
patients with RCC.
The present study aimed to investigate and validate
the value of miR-126, miR-21 and miR-10b expression as prognostic
biomarkers in a Swedish cohort of patients with RCC. An additional
aim was to identify the most suitable endogenous control gene(s)
for miRNA expression studies in ccRCC tissues.
Materials and methods
Study population
Patients were recruited from the Örebro Kidney
Cancer Cohort (OKCC), which consisted of 485 patients consecutively
diagnosed with RCC, who received surgical treatment between January
1, 1986 and December 31, 2013 at the Department of Urology, Örebro
University Hospital, Örebro, Sweden. The patients were followed
through medical records until December 31, 2015 and, in cases where
the patient was deceased, the cause of mortality was established by
the death certificate from the Swedish Cause of Death Register and
classified as mortality from renal cancer or from other causes. The
inclusion criteria for the present study were a diagnosis of ccRCC
and the undergoing of surgery for RCC between 1986 and 2010. Of the
initial 485 patients in the OKCC cohort, 221 patients were excluded
from the study (92 had surgery after 2010, 42 had no tumor sample
available for histological re-evaluation, 6 had benign tumor at
histological re-evaluation, and 81 had other tumor histology than
ccRCC), leaving 264 patients matching the inclusion and exclusion
criteria. Due to the limited amount of tissue available from these
patients, 116 were randomly selected for inclusion in the present
study. Of these 116 patients, 69 had malignant and adjacent benign
tissue available for nucleic acid extraction, and the remaining 47
patients had only malignant tissue available for extraction
(Fig. 1). The selected patient
characteristics are listed in Table
I. The present study was approved by the ethics committee of
the Uppsala and Örebro region, Sweden.
 | Table I.Selected patient demographic and
clinical characteristics (n=116). |
Table I.
Selected patient demographic and
clinical characteristics (n=116).
|
Characteristics | Value |
|---|
| Mean age at
diagnosis (range), years | 66.9 (40–93) |
| Sex, n (%) |
|
|
Male | 62 (53.4) |
|
Female | 54 (46.6) |
| Mean BMI at
diagnosis (range) | 27.1
(18.1–39.5) |
| Smoking status, n
(%) |
|
|
Yes | 35 (30.2) |
| No | 52 (44.8) |
|
Missing | 29 (25.0) |
| Median primary
tumor diameter (range), mm | 70
(20–160) |
| AJCC stage |
|
| I | 49 (42.2) |
| II | 24 (20.7) |
|
III | 26 (22.4) |
| IV | 17 (14.7) |
| Fuhrman grade |
|
| I | 4 (3.4) |
| II | 52 (44.8) |
|
III | 48 (41.4) |
| IV | 12 (10.3) |
| Metastases at
diagnosis |
|
|
Yes | 14
(12.1) |
| No | 102 (87.9) |
| Radical
nephrectomy |
|
|
Yes | 107 (92.2) |
| No | 9
(7.8) |
| Partial
nephrectomy |
|
|
Yes | 10 (8.6) |
| No | 106 (91.4) |
| Recurrence |
|
|
Yes | 49 (42.2) |
| No | 67 (57.8) |
| Cause of
mortality |
|
| Renal
cancer | 48 (51.1) |
| Other
cause | 46 (48.9) |
|
Alive | 22 |
| Median follow-up
time (range), months | 63
(0–302) |
Stability of endogenous control
genes
In order to determine which endogenous control genes
were the most suitable for miRNA expression studies in ccRCC, a
subset of 21 samples were chosen to investigate the stability of 7
previously reported control genes for renal tissues. The samples
were chosen to span across all years (1987–2010) and to include
various tumor stages and grades (Table
II). Malignant and adjacent benign tissues were included in the
investigation (n=42).
 | Table II.Clinicopathological features of
samples included in the selection of endogenous control genes. |
Table II.
Clinicopathological features of
samples included in the selection of endogenous control genes.
| Samplea | Year of
diagnosis | Fuhrman grade | AJCC stage |
|---|
| 1 | 1989 | IV | IV |
| 2 | 1990 | II | I |
| 3 | 1991 | III | IV |
| 4 | 1992 | II | II |
| 5 | 1993 | III | IV |
| 6 | 1994 | II | II |
| 7 | 1995 | II | III |
| 8 | 1996 | III | IV |
| 9 | 1997 | II | I |
| 10 | 1998 | IV | III |
| 11 | 1999 | II | I |
| 12 | 2000 | III | III |
| 13 | 2001 | II | I |
| 14 | 2002 | III | III |
| 15 | 2003 | II | I |
| 16 | 2004 | III | IV |
| 17 | 2005 | II | I |
| 18 | 2006 | III | IV |
| 19 | 2007 | II | IV |
| 20 | 2008 | II | I |
| 21 | 2009 | III | II |
Nucleic acid extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
The patient material in the present study consisted
of formalin-fixed paraffin-embedded (FFPE) tissue obtained from
patients with ccRCC. Malignant and adjacent benign tissue was
marked by two dedicated uro-pathologists (MF and FG) on hematoxylin
and eosin slides corresponding with the paraffin blocks prior to
punching out two cores (1 mm) of tissue from each area. The tissue
cores were deparaffinized according to a standard protocol using
xylene and alcohols prior to the extraction of DNA and total RNA
(including small RNAs) using the AllPrep DNA/RNA FFPE kit (catalog
no. 80234; Qiagen GmbH, Hilden, Germany), according to the
manufacturer's protocol. The quality and quantity of the resulting
DNA and RNA was assessed using the NanoDrop 2000 (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The RNA quality was determined
using the 2100 BioAnalyzer system with the RNA 6000 Pico kit
(Agilent Technologies, Santa Clara, CA, USA). Total RNA samples
were diluted to a concentration of 2 ng/µl prior to RT using the
TaqMan® microRNA reverse transcription kit (catalog no.
4366596; Thermo Fisher Scientific, Inc.) and TaqMan®
assays specific for the miRNAs and endogenous control genes
investigated in the present study. The thermal protocol used for RT
was as follows: 16°C for 30 min, 42°C for 30 and 5 min at 85°C. The
resulting cDNA was mixed with TaqMan® universal master
mix II no AmpErase® UNG (catalog no. 4440040; Thermo
Fisher Scientific, Inc.) and TaqMan® small RNA assays
specific for each miRNA and endogenous control gene (Table III), and used in qPCRs, according
to the manufacturer's protocol. The thermocycling conditions for
qPCR were as follows: 95°C for 10 min, 40 cycles of 95°C for 15 sec
and 60°C for 1 min. All reactions were performed on an Applied
Biosystems 7900HT real-time PCR system and raw Cq values
were determined using SDS software version 2.4 (both Applied
Biosystems; Thermo Fisher Scientific, Inc.). A threshold of 0.2 was
used for each miRNA.
 | Table III.Control genes and miRs investigated
in the present study. |
Table III.
Control genes and miRs investigated
in the present study.
| Namea | TaqMan
assayb | IDc |
|---|
| RNU44 | 001094 | NR_002750 |
| RNU24 | 001001 | NR_002447 |
| RNU48 | 001006 | NR_002745 |
| RNU6B | 001093 | NR_002752 |
| U6 | 001973 | NR_004394 |
| U47 | 001223 | AF141346 |
| RPL21 | 001209 | AB061826 |
| miR-126 | 002228 | hsa-miR-126-3p |
| miR-21 | 000397 | hsa-miR-21-5p |
| miR-10b | 002218 | hsa-miR-10b-5p |
Statistical analysis
The stability of the endogenous control genes was
evaluated using NormFinder (39).
This software application calculates a stability value (SV) defined
as the absolute mean + one standard deviation; which is calculated
using the intergroup variation values from the candidate control
genes. Raw Cq values were used as input. A Wilcoxon
signed rank test was performed for the comparison of the expression
levels of the endogenous control genes between malignant and
adjacent benign samples.
All samples were analyzed in four replicates, and
the mean Cq value was calculated and used in subsequent
analyses. In order to compare the miRNA expression levels between
malignant and adjacent benign tissues, a Wilcoxon test was
performed. The relative up- or downregulation (fold change, FC) of
miRNAs in malignant tissue compared with adjacent benign tissue was
obtained by 2−∆∆Cq method (40), assuming complete efficiency of the
qPCR. A Mann-Whitney U test or Kruskal-Wallis test with the
∆Cq values as input values were subsequently used to
compare miRNA expression levels between groups of patients. The
miRNA expression was divided into high or low expression based on
receiver operating characteristic (ROC) curves with
disease-specific survival as the endpoint. The Kaplan-Meier method
was used to estimate survival probabilities and the log-rank test
was used to compare groups. Cox regression models were used to
estimate hazard ratios (HRs) and 95% confidence intervals (CIs) of
the association between miRNA expression and time to recurrence of
the disease and disease-specific survival. Person-time was
calculated from the date of cancer diagnosis to the earliest of the
following time points: Either mortality from renal cancer or the
recurrence of the disease (depending on the endpoint of the
analysis), censored at time of mortality from other causes, or the
end of follow-up (December 2015). Cox regression models were
performed using miRNAs as dichotomous variables, adjusting for
primary tumor diameter (<40 vs. >40 mm), American Joint
Committee on Cancer (AJCC) stage (I+II vs. III+IV) and Fuhrman
grade (I+II vs. III+IV) (41,42). In
order to evaluate the discriminating capacities of each miRNA,
alone or in combination with other predictive factors, ROC curve
analyses were performed with a calculation of the area under the
curve (AUC) for disease-specific survival. P<0.05 was considered
to indicate a statistically significance. All analyses were
performed in SPSS version 22.0 (IBM Corp., Armonk, NY, USA).
Results
Endogenous control genes
In order to investigate whether the endogenous
control genes used in the present study were stably expressed
across all samples, stability tests were performed on 7 endogenous
control genes (RNU44, RNU24, RNU48, RNU6B, U6, U47 and RPL21) in 21
paired malignant and adjacent benign renal tissue samples. No
significant difference in the expression between malignant and
adjacent benign tissue for any of the control genes tested was
identified (Table II). The
NormFinder software application identified RNU48 (SV, 0.005), U47
(SV, 0.011) and the combination of RNU48 and U47 (SV, 0.006) as the
most stably expressed endogenous control genes within these samples
(Table IV).
 | Table IV.Stability analysis of endogenous
control genes with the NormFinder software. |
Table IV.
Stability analysis of endogenous
control genes with the NormFinder software.
| Control
genea | SV | SE | N |
|---|
| RNU44 | 0.016 | 0.006 | 42 |
| RNU24 | 0.013 | 0.005 | 42 |
| RNU48b | 0.005 | 0.004 | 42 |
| RNU6B | 0.014 | 0.004 | 42 |
| U6 | 0.018 | 0.007 | 42 |
| U47b | 0.01 | 0.005 | 42 |
| RPL21 | 0.016 | 0.006 | 42 |
miRNA expression profiles
differentiate between malignant and adjacent benign tissues
Based on the results from the investigation of
endogenous control genes, the miRNA expression values were
normalized to the geometric mean of RNU48 and U47. The expression
levels of miR-126, miR-21 and miR-10b were investigated in 69
paired samples of malignant and adjacent benign tissues. A
difference in the expression between the malignant and benign
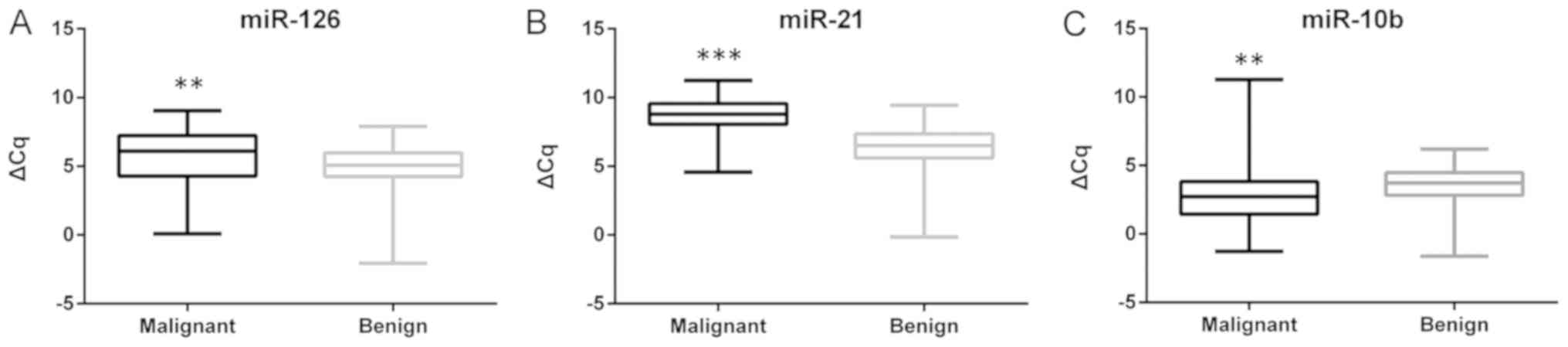
tissues was evident for all three miRNAs, with miR-126 (P=0.004)
and miR-21 (P<0.001) being upregulated, and miR-10b (P<0.001)
being downregulated in malignant tissues compared with adjacent
benign tissue samples (Fig. 2).
Heterogeneity in the up/downregulation of the miRNAs in the samples
was also identified. miR-126 was upregulated in 46 of the 69
(66.7%) malignant samples with a median FC of 3.25 (range,
1.05–66.64). A more consistent upregulation was identified for
miR-21, which was upregulated in 62 of the 69 (89.8%) samples, with
a median FC of 5.95 (range, 1.21–305.57). miR-10b was downregulated
in 48 of the 69 (69.6%) malignant samples, with a median FC of 0.37
(range, 0.01–0.98) in the downregulated samples.
miRNA expression profiles as
prognostic biomarkers
The expression of miR-126, miR-21 and miR-10b was
subsequently investigated in 116 ccRCC tumor specimens from the
OKCC. miR-10b downregulation was associated with increasing AJCC
stage (I+II vs. III+IV; P<0.001) and Fuhrman grade (I+II vs.
III+IV; P<0.001). However, no difference was observed in the
expression of miR-10b in the samples of patients presenting with
metastatic disease at the time of diagnosis compared with that in
patients without metastases (P=0.07). miR-126 downregulation was
also more prominent with increasing AJCC stage (I+II vs. III+IV,
P<0.001) and Fuhrman grade (I+II vs. III+IV, P<0.001), and a
difference in expression was identified in patients with metastases
at the time of diagnosis compared with that in patients without
metastases (P=0.036). The expression of miR-21 was not associated
with any of the clinical variables investigated in the present
study, including AJCC stage, Fuhrman grade or metastatic disease at
the time of diagnosis.
The expression levels of the investigated miRNAs
were subsequently divided into low and high expression groups using
ROC curves. Disease-specific survival time following diagnosis in
the miR-126 low and high groups were 87.8±11.7 and 138.9±17.5
months, respectively (P=0.012; Fig.
3A). Patients with low miR-126 expression also had shorter time
to recurrence of disease, compared with that of patients with high
miR-126 expression, 124.9±17.9 and 155.4±13.9 months, respectively
(P=0.007; Fig. 3B). No differences
in the time to recurrence or disease-specific survival were
identified between the low and high miR-10b or miR-21 expression
groups (P>0.05).
In the multivariate analysis, the patients with low
miR-126 expression tended to exhibit a shorter recurrence time
compared with that of patients with high expression (adjusted HR,
1.79; 95% CI, 0.91–3.52). Patients with low miR-126 expression also
tended to exhibit shorter disease-specific survival time (adjusted
HR, 1.94; 95% CI, 0.95–3.97) (Table
V). Neither miR-10b nor miR-21 expression (low versus high) was
associated with recurrence or disease-specific survival times.
 | Table V.Cox regression analysis of low versus
high miR-126 expression. |
Table V.
Cox regression analysis of low versus
high miR-126 expression.
|
| HR (95% CI) |
|---|
|
|
|
|---|
| Parameter | Crude model | Adjusted
modela |
|---|
| Disease
recurrence | 2.32
(1.23–4.38) | 1.79
(0.91–3.52) |
| Disease-specific
survival | 2.32
(1.18–4.55) | 1.94
(0.95–3.97) |
In order to explore the accuracy of miRNA expression
and the presently used prognostic factors (Fuhrman grade, AJCC
stage and primary tumor diameter) as classifiers of lethal renal
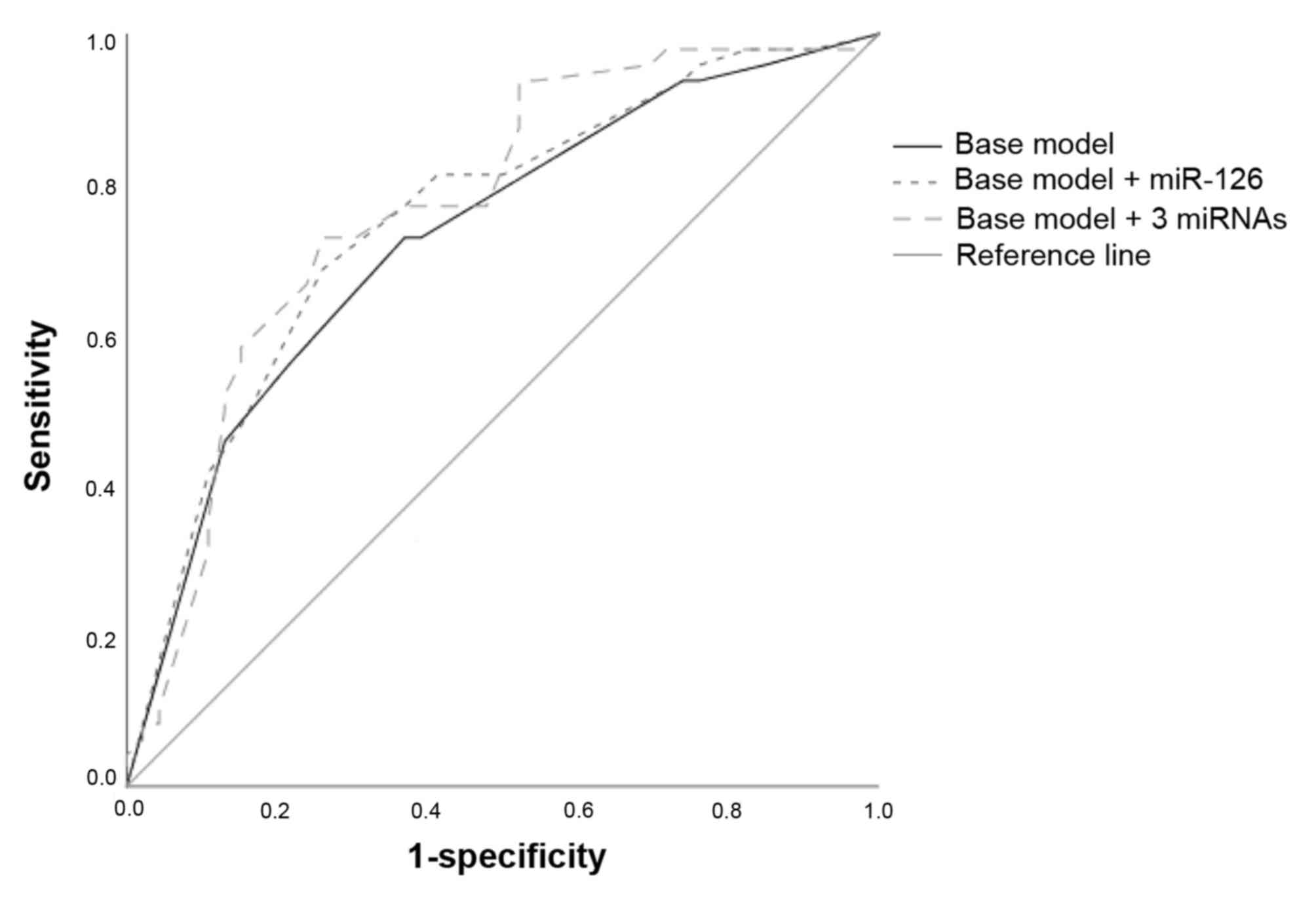
cancer, ROC curves were employed. The baseline model, including
Fuhrman grade, AJCC stage and primary tumor diameter, yielded an
AUC of 0.73 (95% CI, 0.63–0.83). Adding the dichotomized (low
versus high) expression of miR-126 yielded an insignificant
increase of the AUC (AUC, 0.754; 95% CI, 0.66–0.85; P>0.05).
Including the dichotomized expression of all three miRNAs further
increased the AUC (AUC, 0.77; 95% CI, 0.67–0.87; P>0.05;
Fig. 4); however, this increase was
not statistically significant. No single threshold for any
measurement of the combined model yielded high sensitivity combined
with high specificity.
Subgroup analysis
In order to investigate whether the expression of
miR-126, miR-21 or miR-10b may be used as reliable biomarkers for
aggressive disease in patients with a primary tumor diameter of ≤40
mm, a subgroup analysis was performed. Patients with a primary
tumor diameter of >40 mm and low miR-126 expression exhibited
shorter recurrence time (P=0.035) and disease-specific survival
time (P=0.028), compared with patients with high expression.
However, no difference in the time to disease recurrence or
disease-specific mortality were identified between patients with
low or high miR-126 expression who had a primary tumor diameter
<40 mm. When adjusting for other clinical parameters, no
association between miR-126 expression and time to recurrence or
disease-specific mortality was observed in patients with a primary
tumor diameter of <40 nor >40 mm.
Discussion
Previous studies have investigated the potential
role of miRNAs as biomarkers for RCC (14,15,17-21,23,43-50).
However, the value of using miRNAs as prognostic biomarkers for RCC
is still under debate. The present study selected the miRNAs
miR-126, miR-21, and miR-10b, which have previously been suggested
as prognostic biomarkers for RCC, and aimed to validate their
usefulness as prognostic markers in a Swedish cohort of patients
with RCC. The results of the present study demonstrated that all
three miRNAs were deregulated in malignant tissues when compared
with adjacent benign tissues. However, the present study could only
confirm a potential prognostic value for the expression of miR-126,
but not for that of miR-21 or miR-10b.
In order to minimize the influence of technical
variation and to increase the expression accuracy of biological
data, the normalization of gene expression data is required. Due to
the low number of target genes investigated, population-based
normalization methods commonly used in microarray studies are not
suitable for qPCR data, and normalization is instead dependent on
the use of endogenous control genes as references for normal
expression (51). No universal
control gene suitable for all experiments exists, and it has
previously been recommended to test the most suitable control gene
specific for the experiment performed (52). A total of seven candidate control
genes for miRNA expression studies in FFPE tissues from patients
with ccRCC were investigated in the present study. The results
indicate that RNU48, followed by U47, or the combination of these
genes, were the most suitable for normalization of miRNA expression
within the present study. Since normalization based on a single
control gene could lead to erroneous results, the use of multiple
control genes for normalization is preferred (52,53).
Therefore, the normalization of the RT-qPCR data in the present
study was performed using the geometric mean of RNU48 and U47
expression.
The deregulation of miR-126 has been identified in a
variety of cancer types; however, its role in carcinogenesis
remains to be elucidated. The expression of miR-126 has been
observed to be downregulated in lung and esophageal cancer
(27,28). On the other hand, the same miRNA has
been identified to be upregulated in urothelial and gastric cancer
(29,30). In RCC, the expression of miR-126 has
been reported to serve a role in the molecular classification of
histological subtypes (54). The
results of the present study demonstrate that miR-126 was
upregulated in ccRCC, and that the expression of miR-126 was
decreased with increasing grade and tumor stage, which is
consistent with previous findings for RCC (15,19,20,49).
However, the present study observed a heterogeneity of miR-126
expression in malignant samples, being upregulated in 66.7% and
downregulated in 33.3% of the investigated samples. Similar
findings were observed by Vergho et al (15), who hypothesized that miR-126 was
up/downregulated in different subgroups of patients with RCC. The
same authors later reported a downregulation of miR-126 in ccRCC
cases with tumor thrombus of the inferior vena cava, but not in
cases without tumor thrombus (20).
These results suggest that the downregulation of miR-126 in the
primary tumor is associated with a more invasive phenotype. This
hypothesis is supported by the data of others including those of
the present study, demonstrating that patients with metastatic
disease at the time of diagnosis have lower miR-126 expression in
the primary tumor compared with patients presenting without
metastases at diagnosis (15,19,20,48,49).
A low miR-126 expression was also associated with early recurrence
and shorter disease-specific survival time in the univariate, but
not multivariate, analyses, which was also consistent with results
obtained from previous studies (15,20,47). A
ROC analysis indicated that the addition of miR-126 expression to
currently used clinical factors could improve the prognostic
accuracy. However, this small increase in AUC may not easily
translate to the clinical setting.
An early event in the pathogenesis of RCC is the
inactivation of the tumor suppressor Von Hippel-Lindau
(VHL), which is a direct target gene of miR-21 (55). As the downregulation of VHL
may be caused by the upregulation of miR-21, the deregulation of
miR-21 could constitute an early event in renal carcinogenesis.
Several studies have demonstrated that miR-21 is overexpressed in
renal tumors compared with benign renal tissue (14,15,20,21,44,46,49,50). In
the present study, miR-21 was identified to be upregulated in 89.8%
of the ccRCC tissues, in line with a study performed by Zaman et
al (21), who reported miR-21
upregulation in 89% of the investigated renal tumors. Previous
studies have also observed an increased expression of miR-21 in
increasing grades and stages of RCC (14,15), and
that patients with metastatic disease at the time of diagnosis had
a higher expression of miR-21 in their primary tumor (14,15,20,21,49).
However, these results could not be confirmed in the present study,
as no association between miR-21 expression and tumor grade, stage
or patient outcome was identified.
miR-10b is considered to be associated with the
metastatic behavior of tumors, partly due to its role as a
suppressor of homeobox D10, a transcriptional repressor that
inhibits expression of genes involved in cell migration and
extracellular matrix remodeling (56). In the present study, miR-10b was
observed to be downregulated in ccRCC tissues compared with
adjacent benign tissue, a finding supported by previous studies
(18,44,46,49).
This investigation did not reveal a difference in the expression of
miR-10b between patients presenting with or without metastatic
disease at the time of diagnosis. However, several studies have
reported miR-10b as one of the most downregulated miRNAs in
metastatic tissue, with a gradual downregulation of the expression
between normal tissue, primary tumor and metastatic tissue
(43,48,49).
Patients with a high expression of miR-10b have previously been
demonstrated to have longer progression-free and disease-free
survival times (18,42). This could not be confirmed in the
present study, as no association between miR-10b expression and
clinical outcome was evident.
The discrepancies between the results of the present
study and previous studies may be due to a number of factors,
including differences in patient populations, timing and method of
tissue sampling, and the use of fresh-frozen or FFPE tissue.
Furthermore, the analysis method (microarray, qPCR or next
generation sequencing), or choice of endogenous control genes for
normalization of qPCR data may also influence the results of a
study. A strength of the present study is the investigation of
endogenous control genes within the cohort. Numerous studies use
constitutively expressed housekeeping genes for normalization,
without proper validation of their presumed stability of
expression, which may produce erroneous results. If miRNAs are to
be used as biomarkers for RCC, a standardized analysis scheme
should be optimized for all these variables.
Another strength of the present study is the
well-defined cohort used, including complete follow-up of the study
participants, with well defined outcome measurements from the
Swedish Cause of Death register. Many of the previously performed
studies have limitations due to relatively short follow-up periods
of the patients. A caveat within this study is that due to the
relatively small study population, there was not enough statistical
power to investigate the prognostic potential of miRNA expression
in subgroups of patients (e.g., metastatic versus non-metastatic
disease). Therefore, additional studies are required to further
explore the prognostic potential of miRNA expression in subgroups
of patients with ccRCC. Furthermore, only three of almost 2,000
known miRNAs were investigated in the present study, which limits
the opportunity to identify novel potential prognostic biomarkers
for ccRCC.
To conclude, several prognostic factors for RCC are
presently being used in a clinical setting; however, these factors
lack accuracy in predicting the natural history of the disease,
particularly in patients with non-metastatic disease at the time of
diagnosis. Therefore, the identification of biomarkers that can aid
in predicting patient outcome for patients with RCC, either alone
or in combination with currently used clinical parameters are
required. The results of the present study confirm that all of the
investigated miRNAs are deregulated in malignant tissue compared
with adjacent benign tissue. However, only one of the miRNAs
investigated, miR-126, exhibited prognostic potential among the
included patients.
Acknowledgements
The authors would like to acknowledge Dr Beata
Grabowska (Department of Urology, University Hospital of Örebro,
Sweden) who helped obtain the clinical information of the patients
included in the study.
Funding
This study was supported by the Örebro county
research council (grant no. OLL-430521). The funding source had no
role in the design of the study, data collection, analyses,
interpretation of data or manuscript writing.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JCa, SD, PS, MF, and FG designed the study. MF and
FG performed all histological analyses. JCa and JCh performed the
laboratory and statistical analyses. JCa, JC, SD, and PS
interpreted the results. All authors helped to draft the manuscript
and approved the final version.
Ethics approval and consent to
participate
The present study was approved by the ethics
committee of the Uppsala and Örebro region (reference number
2015/353). The ethics committee considered that no consent from
patients included in the study was needed.
Patient consent for publication
The ethics committee considered that no consent from
patients included in the study was needed since all data are
presented on a group level and the published results cannot be
traced back to an individual patient.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lopez-Beltran A, Carrasco JC, Cheng L,
Scarpelli M, Kirkali Z and Montironi R: 2009 update on the
classification of renal epithelial tumors in adults. Int J Urol.
16:432–443. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ljungberg B, Alamdari FI, Rasmuson T and
Roos G: Follow-up guidelines of nonmetastatic renal cell carcinoma
based on the occurrence of metastases after radical nephrectomy.
BJU Int. 84:405–411. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Regional Cancer Center: Renal
cancer-National quality register report of 2015. https://www.cancercentrum.se/globalassets/cancerdiagnoser/urinvagar/njurcancer/natvp_njurcancer_2017-09-13_final.pdf?v=c90d6e26233847b9801df004ceb97ae2October
13–2017(In Swedish).
|
|
5
|
Diaz de Leon A and Pedrosa I: Imaging and
screening of kidney cancer. Radiol Clin North Am. 55:1235–1250.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Azawi NH, Lund L and Fode M: Renal cell
carcinomas mass of <4 cm are not always indolent. Urol Ann.
9:234–238. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Delahunt B, Srigley JR, Montironi R and
Egevad L: Advances in renal neoplasia: Recommendations from the
2012 international society of urological pathology consensus
conference. Urology. 83:969–974. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nerich V, Hugues M, Paillard MJ, Borowski
L, Nai T, Stein U, Nguyen Tan Hon T, Montcuquet P, Maurina T,
Mouillet G, et al: Clinical impact of targeted therapies in
patients with metastatic clear-cell renal cell carcinoma. Onco
Targets Ther. 7:365–374. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Heng DY, Xie W, Regan MM, Harshman LC,
Bjarnason GA, Vaishampayan UN, Mackenzie M, Wood L, Donskov F, Tan
MH, et al: External validation and comparison with other models of
the international metastatic renal-cell carcinoma database
consortium prognostic model: A population-based study. Lancet
Oncol. 14:141–148. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Backes C, Meese E, Lenhof HP and Keller A:
A dictionary on microRNAs and their putative target pathways.
Nucleic Acids Res. 38:4476–4486. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Faragalla H, Youssef YM, Scorilas A,
Khalil B, White NM, Mejia-Guerrero S, Khella H, Jewett MA, Evans A,
Lichner Z, et al: The clinical utility of miR-21 as a diagnostic
and prognostic marker for renal cell carcinoma. J Mol Diagn.
14:385–392. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vergho D, Kneitz S, Rosenwald A, Scherer
C, Spahn M, Burger M, Riedmiller H and Kneitz B: Combination of
expression levels of miR-21 and miR-126 is associated with
cancer-specific survival in clear-cell renal cell carcinoma. BMC
Cancer. 14:252014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fritz HKM, Lindgren D, Ljungberg B,
Axelson H and Dahlback B: The miR(21/10b) ratio as a prognostic
marker in clear cell renal cell carcinoma. Eur J Cancer.
50:1758–1765. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He C, Zhao X, Jiang H, Zhong Z and Xu R:
Demethylation of miR-10b plays a suppressive role in ccRCC cells.
Int J Clin Exp Pathol. 8:10595–10604. 2015.PubMed/NCBI
|
|
18
|
Khella HWZ, Daniel N, Youssef L, Scorilas
A, Nofech-Mozes R, Mirham L, Krylov SN, Liandeau E, Krizova A,
Finelli A, et al: miR-10b is a prognostic marker in clear cell
renal cell carcinoma. J Clin Pathol. 70:854–859. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Khella HW, Scorilas A, Mozes R, Mirham L,
Lianidou E, Krylov SN, Lee JY, Ordon M, Stewart R, Jewett MA and
Yousef GM: Low expression of miR-126 is a prognostic marker for
metastatic clear cell renal cell carcinoma. Am J Pathol.
185:693–703. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vergho DC, Kneitz S, Kalogirou C, Burger
M, Krebs M, Rosenwald A, Spahn M, Löser A, Kocot A, Riedmiller H
and Kneitz B: Impact of miR-21, miR-126 and miR-221 as prognostic
factors of clear cell renal cell carcinoma with tumor thrombus of
the inferior vena cava. PLoS One. 9:e1098772014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zaman MS, Shahryari V, Deng G, Thamminana
S, Saini S, Majid S, Chang I, Hirata H, Ueno K, Yamamura S, et al:
Up-regulation of microRNA-21 correlates with lower kidney cancer
survival. PLoS One. 7:e310602012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu Z, Liu M, Stribinskis V, Klinge CM,
Ramos KS, Colburn NH and Li Y: MicroRNA-21 promotes cell
transformation by targeting the programmed cell death 4 gene.
Oncogene. 27:4373–4379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang A, Liu Y, Shen Y, Xu Y and Li X:
miR-21 modulates cell apoptosis by targeting multiple genes in
renal cell carcinoma. Urology. 78:474 e413–479. 2011. View Article : Google Scholar
|
|
24
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu SG, Qin XG, Zhao BS, Qi B, Yao WJ,
Wang TY, Li HC and Wu XN: Differential expression of miRNAs in
esophageal cancer tissue. Oncol Lett. 5:1639–1642. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guan P, Yin Z, Li X, Wu W and Zhou B:
Meta-analysis of human lung cancer microRNA expression profiling
studies comparing cancer tissues with normal tissues. J Exp Clin
Cancer Res. 31:542012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Snowdon J, Boag S, Feilotter H, Izard J
and Siemens DR: A pilot study of urinary microRNA as a biomarker
for urothelial cancer. Can Urol Assoc J. 7:28–32. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Otsubo T, Akiyama Y, Hashimoto Y, Shimada
S, Goto K and Yuasa Y: MicroRNA-126 inhibits SOX2 expression and
contributes to gastric carcinogenesis. PLoS One. 6:e166172011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu B, Peng XC, Zheng XL, Wang J and Qin
YW: MiR-126 restoration down-regulate VEGF and inhibit the growth
of lung cancer cell lines in vitro and in vivo. Lung Cancer.
66:169–175. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu
T, Bai Y, Shen Y, Yuan W, Jing Q and Qin Y: Endothelial-specific
intron-derived miR-126 is down-regulated in human breast cancer and
targets both VEGFA and PIK3R2. Mol Cell Biochem. 351:157–164. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo C, Sah JF, Beard L, Willson JK,
Markowitz SD and Guda K: The noncoding RNA, miR-126, suppresses the
growth of neoplastic cells by targeting phosphatidylinositol
3-kinase signaling and is frequently lost in colon cancers. Genes
Chromosomes Cancer. 47:939–946. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Harris TA, Yamakuchi M, Ferlito M, Mendell
JT and Lowenstein CJ: MicroRNA-126 regulates endothelial expression
of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA.
105:1516–1521. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ahmad A, Sethi S, Chen W, Ali-Fehmi R,
Mittal S and Sarkar FH: Up-regulation of microRNA-10b is associated
with the development of breast cancer brain metastasis. Am J Transl
Res. 6:384–390. 2014.PubMed/NCBI
|
|
36
|
Wang YY, Li L, Ye ZY, Zhao ZS and Yan ZL:
MicroRNA-10b promotes migration and invasion through Hoxd10 in
human gastric cancer. World J Surg Oncol. 13:2592015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun L, Yan W, Wang Y, Sun G, Luo H, Zhang
J, Wang X, You Y, Yang Z and Liu N: MicroRNA-10b induces glioma
cell invasion by modulating MMP-14 and uPAR expression via HOXD10.
Brain Res. 1389:9–18. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Y, Chen D, Jin L, Liu J, Su Z, Qi Z,
Shi M, Jiang Z, Ni L, Yang S, et al: Oncogenic cAMP responsive
element binding protein 1 is overexpressed upon loss of tumor
suppressive miR-10b-5p and miR-363-3p in renal cancer. Oncol Rep.
35:1967–1978. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Andersen CL, Jensen JL and Orntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: A model-based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Cancer Res. 64:5245–5250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Edge SB, Byrd D, Compton C, Fritz AG,
Greene FL and Trotti A III: AJCC cancer staging manual (7th).
Springer-Verlag. New York, NY: 2010.
|
|
42
|
Fuhrman SA, Lasky LC and Limas C:
Prognostic significance of morphologic parameters in renal cell
carcinoma. Am J Surg Pathol. 6:655–663. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Heinzelmann J, Henning B, Sanjmyatav J,
Posorski N, Steiner T, Wunderlich H, Gajda MR and Junker K:
Specific miRNA signatures are associated with metastasis and poor
prognosis in clear cell renal cell carcinoma. World J Urol.
29:367–373. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Juan D, Alexe G, Antes T, Liu H,
Madabhushi A, Delisi C, Ganesan S, Bhanot G and Liou LS:
Identification of a microRNA panel for clear-cell kidney cancer.
Urology. 75:835–841. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jung M, Mollenkopf HJ, Grimm C, Wagner I,
Albrecht M, Waller T, Pilarsky C, Johannsen M, Stephan C, Lehrach
H, et al: MicroRNA profiling of clear cell renal cell cancer
identifies a robust signature to define renal malignancy. J Cell
Mol Med. 13:3918–3928. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Osanto S, Qin Y, Buermans HP, Berkers J,
Lerut E, Goeman JJ and van Poppel H: Genome-wide microRNA
expression analysis of clear cell renal cell carcinoma by next
generation deep sequencing. PLoS One. 7:e382982012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Slaby O, Redova M, Poprach A, Nekvindova
J, Iliev R, Radova L, Lakomy R, Svoboda M and Vyzula R:
Identification of MicroRNAs associated with early relapse after
nephrectomy in renal cell carcinoma patients. Genes Chromosomes
Cancer. 51:707–716. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
White NM, Khella HW, Grigull J, Adzovic S,
Youssef YM, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Jewett MA,
et al: miRNA profiling in metastatic renal cell carcinoma reveals a
tumour-suppressor effect for miR-215. Br J Cancer. 105:1741–1749.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wotschofsky Z, Liep J, Meyer HA, Jung M,
Wagner I, Disch AC, Schaser KD, Melcher I, Kilic E, Busch J, et al:
Identification of metastamirs as metastasis-associated microRNAs in
clear cell renal cell carcinomas. Int J Biol Sci. 8:1363–1374.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yi Z, Fu Y, Zhao S, Zhang X and Ma C:
Differential expression of miRNA patterns in renal cell carcinoma
and nontumorous tissues. J Cancer Res Clin Oncol. 136:855–862.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Peltier HJ and Latham GJ: Normalization of
microRNA expression levels in quantitative RT-PCR assays:
Identification of suitable reference RNA targets in normal and
cancerous human solid tissues. RNA. 14:844–852. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by geometric
averaging of multiple internal control genes. Genome Biol.
3:RESEARCH00342002. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tricarico C, Pinzani P, Bianchi S,
Paglierani M, Distante V, Pazzagli M, Bustin SA and Orlando C:
Quantitative real-time reverse transcription polymerase chain
reaction: Normalization to rRNA or single housekeeping genes is
inappropriate for human tissue biopsies. Anal Biochem. 309:293–300.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Youssef YM, White NM, Grigull J, Krizova
A, Samy C, Mejia-Guerrero S, Evans A and Yousef GM: Accurate
molecular classification of kidney cancer subtypes using microRNA
signature. Eur Urol. 59:721–730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang KL, Han L, Chen LY, Shi ZD, Yang M,
Ren Y, Chen LC, Zhang JX, Pu PY and Kang CS: Blockage of a
miR-21/EGFR regulatory feedback loop augments anti-EGFR therapy in
glioblastomas. Cancer Lett. 342:139–149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|