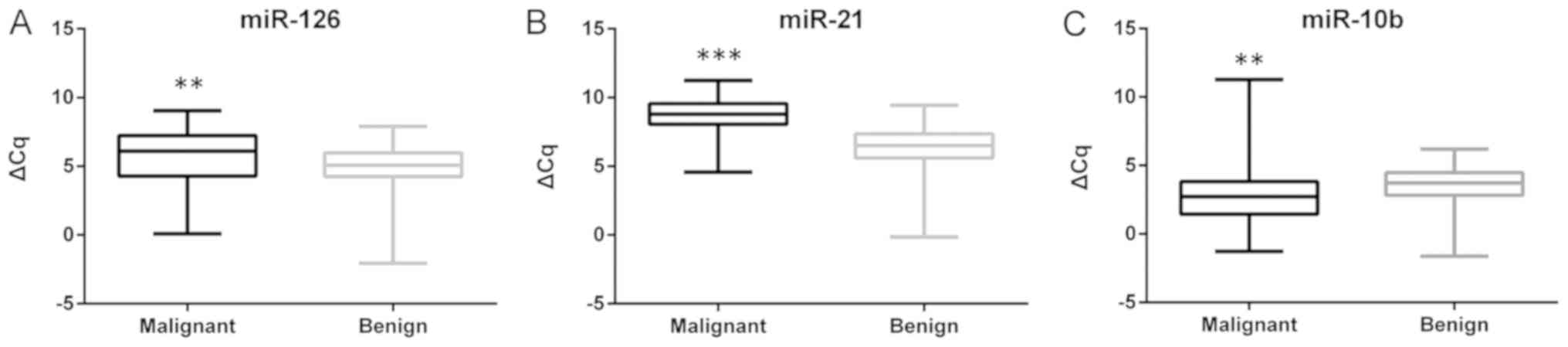
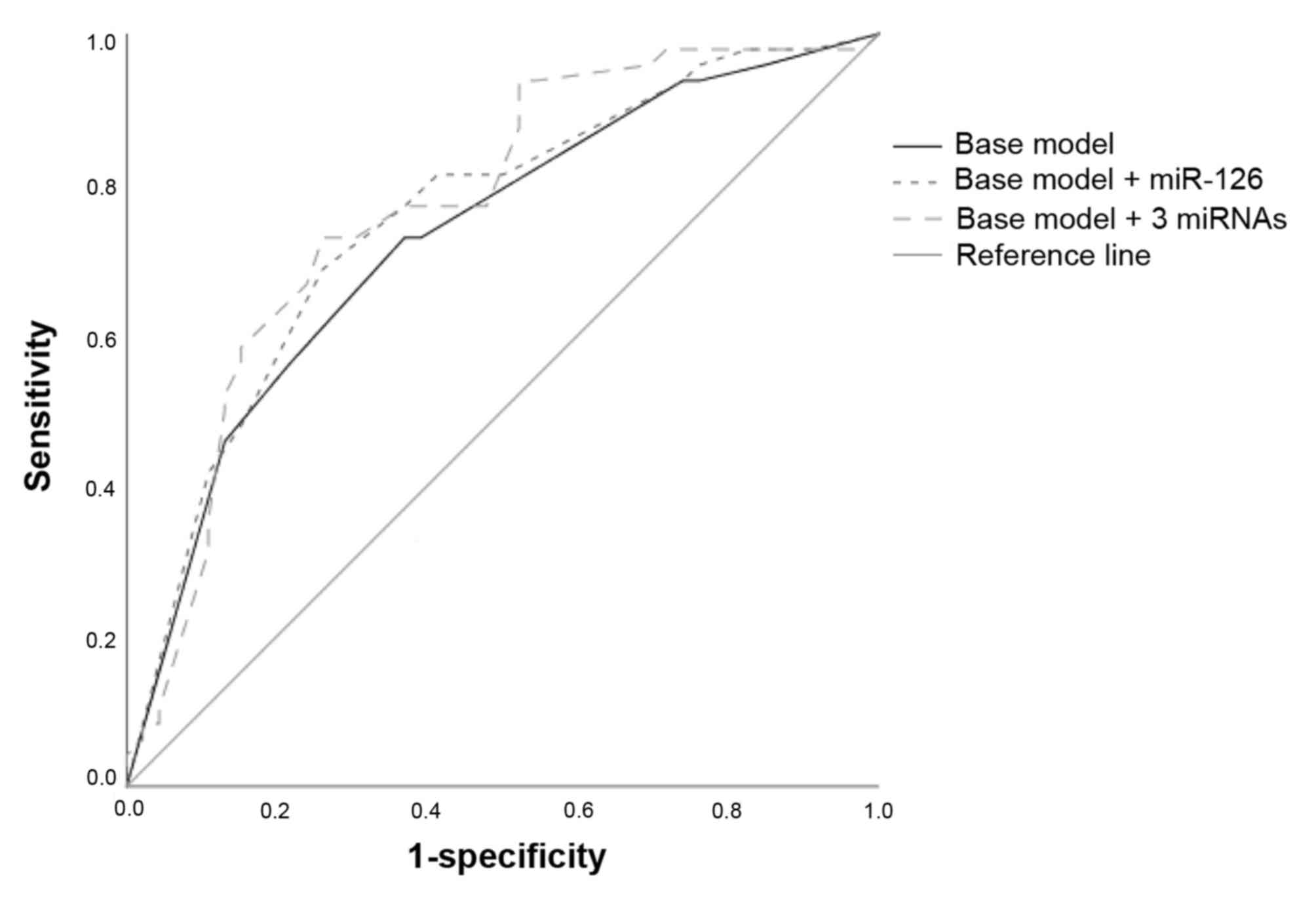
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lopez-Beltran A, Carrasco JC, Cheng L,
Scarpelli M, Kirkali Z and Montironi R: 2009 update on the
classification of renal epithelial tumors in adults. Int J Urol.
16:432–443. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ljungberg B, Alamdari FI, Rasmuson T and
Roos G: Follow-up guidelines of nonmetastatic renal cell carcinoma
based on the occurrence of metastases after radical nephrectomy.
BJU Int. 84:405–411. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Regional Cancer Center: Renal
cancer-National quality register report of 2015. https://www.cancercentrum.se/globalassets/cancerdiagnoser/urinvagar/njurcancer/natvp_njurcancer_2017-09-13_final.pdf?v=c90d6e26233847b9801df004ceb97ae2October
13–2017(In Swedish).
|
|
5
|
Diaz de Leon A and Pedrosa I: Imaging and
screening of kidney cancer. Radiol Clin North Am. 55:1235–1250.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Azawi NH, Lund L and Fode M: Renal cell
carcinomas mass of <4 cm are not always indolent. Urol Ann.
9:234–238. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Delahunt B, Srigley JR, Montironi R and
Egevad L: Advances in renal neoplasia: Recommendations from the
2012 international society of urological pathology consensus
conference. Urology. 83:969–974. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nerich V, Hugues M, Paillard MJ, Borowski
L, Nai T, Stein U, Nguyen Tan Hon T, Montcuquet P, Maurina T,
Mouillet G, et al: Clinical impact of targeted therapies in
patients with metastatic clear-cell renal cell carcinoma. Onco
Targets Ther. 7:365–374. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Heng DY, Xie W, Regan MM, Harshman LC,
Bjarnason GA, Vaishampayan UN, Mackenzie M, Wood L, Donskov F, Tan
MH, et al: External validation and comparison with other models of
the international metastatic renal-cell carcinoma database
consortium prognostic model: A population-based study. Lancet
Oncol. 14:141–148. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Backes C, Meese E, Lenhof HP and Keller A:
A dictionary on microRNAs and their putative target pathways.
Nucleic Acids Res. 38:4476–4486. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Faragalla H, Youssef YM, Scorilas A,
Khalil B, White NM, Mejia-Guerrero S, Khella H, Jewett MA, Evans A,
Lichner Z, et al: The clinical utility of miR-21 as a diagnostic
and prognostic marker for renal cell carcinoma. J Mol Diagn.
14:385–392. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vergho D, Kneitz S, Rosenwald A, Scherer
C, Spahn M, Burger M, Riedmiller H and Kneitz B: Combination of
expression levels of miR-21 and miR-126 is associated with
cancer-specific survival in clear-cell renal cell carcinoma. BMC
Cancer. 14:252014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fritz HKM, Lindgren D, Ljungberg B,
Axelson H and Dahlback B: The miR(21/10b) ratio as a prognostic
marker in clear cell renal cell carcinoma. Eur J Cancer.
50:1758–1765. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He C, Zhao X, Jiang H, Zhong Z and Xu R:
Demethylation of miR-10b plays a suppressive role in ccRCC cells.
Int J Clin Exp Pathol. 8:10595–10604. 2015.PubMed/NCBI
|
|
18
|
Khella HWZ, Daniel N, Youssef L, Scorilas
A, Nofech-Mozes R, Mirham L, Krylov SN, Liandeau E, Krizova A,
Finelli A, et al: miR-10b is a prognostic marker in clear cell
renal cell carcinoma. J Clin Pathol. 70:854–859. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Khella HW, Scorilas A, Mozes R, Mirham L,
Lianidou E, Krylov SN, Lee JY, Ordon M, Stewart R, Jewett MA and
Yousef GM: Low expression of miR-126 is a prognostic marker for
metastatic clear cell renal cell carcinoma. Am J Pathol.
185:693–703. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vergho DC, Kneitz S, Kalogirou C, Burger
M, Krebs M, Rosenwald A, Spahn M, Löser A, Kocot A, Riedmiller H
and Kneitz B: Impact of miR-21, miR-126 and miR-221 as prognostic
factors of clear cell renal cell carcinoma with tumor thrombus of
the inferior vena cava. PLoS One. 9:e1098772014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zaman MS, Shahryari V, Deng G, Thamminana
S, Saini S, Majid S, Chang I, Hirata H, Ueno K, Yamamura S, et al:
Up-regulation of microRNA-21 correlates with lower kidney cancer
survival. PLoS One. 7:e310602012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu Z, Liu M, Stribinskis V, Klinge CM,
Ramos KS, Colburn NH and Li Y: MicroRNA-21 promotes cell
transformation by targeting the programmed cell death 4 gene.
Oncogene. 27:4373–4379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang A, Liu Y, Shen Y, Xu Y and Li X:
miR-21 modulates cell apoptosis by targeting multiple genes in
renal cell carcinoma. Urology. 78:474 e413–479. 2011. View Article : Google Scholar
|
|
24
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu SG, Qin XG, Zhao BS, Qi B, Yao WJ,
Wang TY, Li HC and Wu XN: Differential expression of miRNAs in
esophageal cancer tissue. Oncol Lett. 5:1639–1642. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guan P, Yin Z, Li X, Wu W and Zhou B:
Meta-analysis of human lung cancer microRNA expression profiling
studies comparing cancer tissues with normal tissues. J Exp Clin
Cancer Res. 31:542012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Snowdon J, Boag S, Feilotter H, Izard J
and Siemens DR: A pilot study of urinary microRNA as a biomarker
for urothelial cancer. Can Urol Assoc J. 7:28–32. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Otsubo T, Akiyama Y, Hashimoto Y, Shimada
S, Goto K and Yuasa Y: MicroRNA-126 inhibits SOX2 expression and
contributes to gastric carcinogenesis. PLoS One. 6:e166172011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu B, Peng XC, Zheng XL, Wang J and Qin
YW: MiR-126 restoration down-regulate VEGF and inhibit the growth
of lung cancer cell lines in vitro and in vivo. Lung Cancer.
66:169–175. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu
T, Bai Y, Shen Y, Yuan W, Jing Q and Qin Y: Endothelial-specific
intron-derived miR-126 is down-regulated in human breast cancer and
targets both VEGFA and PIK3R2. Mol Cell Biochem. 351:157–164. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo C, Sah JF, Beard L, Willson JK,
Markowitz SD and Guda K: The noncoding RNA, miR-126, suppresses the
growth of neoplastic cells by targeting phosphatidylinositol
3-kinase signaling and is frequently lost in colon cancers. Genes
Chromosomes Cancer. 47:939–946. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Harris TA, Yamakuchi M, Ferlito M, Mendell
JT and Lowenstein CJ: MicroRNA-126 regulates endothelial expression
of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA.
105:1516–1521. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ahmad A, Sethi S, Chen W, Ali-Fehmi R,
Mittal S and Sarkar FH: Up-regulation of microRNA-10b is associated
with the development of breast cancer brain metastasis. Am J Transl
Res. 6:384–390. 2014.PubMed/NCBI
|
|
36
|
Wang YY, Li L, Ye ZY, Zhao ZS and Yan ZL:
MicroRNA-10b promotes migration and invasion through Hoxd10 in
human gastric cancer. World J Surg Oncol. 13:2592015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun L, Yan W, Wang Y, Sun G, Luo H, Zhang
J, Wang X, You Y, Yang Z and Liu N: MicroRNA-10b induces glioma
cell invasion by modulating MMP-14 and uPAR expression via HOXD10.
Brain Res. 1389:9–18. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Y, Chen D, Jin L, Liu J, Su Z, Qi Z,
Shi M, Jiang Z, Ni L, Yang S, et al: Oncogenic cAMP responsive
element binding protein 1 is overexpressed upon loss of tumor
suppressive miR-10b-5p and miR-363-3p in renal cancer. Oncol Rep.
35:1967–1978. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Andersen CL, Jensen JL and Orntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: A model-based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Cancer Res. 64:5245–5250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Edge SB, Byrd D, Compton C, Fritz AG,
Greene FL and Trotti A III: AJCC cancer staging manual (7th).
Springer-Verlag. New York, NY: 2010.
|
|
42
|
Fuhrman SA, Lasky LC and Limas C:
Prognostic significance of morphologic parameters in renal cell
carcinoma. Am J Surg Pathol. 6:655–663. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Heinzelmann J, Henning B, Sanjmyatav J,
Posorski N, Steiner T, Wunderlich H, Gajda MR and Junker K:
Specific miRNA signatures are associated with metastasis and poor
prognosis in clear cell renal cell carcinoma. World J Urol.
29:367–373. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Juan D, Alexe G, Antes T, Liu H,
Madabhushi A, Delisi C, Ganesan S, Bhanot G and Liou LS:
Identification of a microRNA panel for clear-cell kidney cancer.
Urology. 75:835–841. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jung M, Mollenkopf HJ, Grimm C, Wagner I,
Albrecht M, Waller T, Pilarsky C, Johannsen M, Stephan C, Lehrach
H, et al: MicroRNA profiling of clear cell renal cell cancer
identifies a robust signature to define renal malignancy. J Cell
Mol Med. 13:3918–3928. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Osanto S, Qin Y, Buermans HP, Berkers J,
Lerut E, Goeman JJ and van Poppel H: Genome-wide microRNA
expression analysis of clear cell renal cell carcinoma by next
generation deep sequencing. PLoS One. 7:e382982012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Slaby O, Redova M, Poprach A, Nekvindova
J, Iliev R, Radova L, Lakomy R, Svoboda M and Vyzula R:
Identification of MicroRNAs associated with early relapse after
nephrectomy in renal cell carcinoma patients. Genes Chromosomes
Cancer. 51:707–716. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
White NM, Khella HW, Grigull J, Adzovic S,
Youssef YM, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Jewett MA,
et al: miRNA profiling in metastatic renal cell carcinoma reveals a
tumour-suppressor effect for miR-215. Br J Cancer. 105:1741–1749.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wotschofsky Z, Liep J, Meyer HA, Jung M,
Wagner I, Disch AC, Schaser KD, Melcher I, Kilic E, Busch J, et al:
Identification of metastamirs as metastasis-associated microRNAs in
clear cell renal cell carcinomas. Int J Biol Sci. 8:1363–1374.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yi Z, Fu Y, Zhao S, Zhang X and Ma C:
Differential expression of miRNA patterns in renal cell carcinoma
and nontumorous tissues. J Cancer Res Clin Oncol. 136:855–862.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Peltier HJ and Latham GJ: Normalization of
microRNA expression levels in quantitative RT-PCR assays:
Identification of suitable reference RNA targets in normal and
cancerous human solid tissues. RNA. 14:844–852. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by geometric
averaging of multiple internal control genes. Genome Biol.
3:RESEARCH00342002. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tricarico C, Pinzani P, Bianchi S,
Paglierani M, Distante V, Pazzagli M, Bustin SA and Orlando C:
Quantitative real-time reverse transcription polymerase chain
reaction: Normalization to rRNA or single housekeeping genes is
inappropriate for human tissue biopsies. Anal Biochem. 309:293–300.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Youssef YM, White NM, Grigull J, Krizova
A, Samy C, Mejia-Guerrero S, Evans A and Yousef GM: Accurate
molecular classification of kidney cancer subtypes using microRNA
signature. Eur Urol. 59:721–730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang KL, Han L, Chen LY, Shi ZD, Yang M,
Ren Y, Chen LC, Zhang JX, Pu PY and Kang CS: Blockage of a
miR-21/EGFR regulatory feedback loop augments anti-EGFR therapy in
glioblastomas. Cancer Lett. 342:139–149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|