Introduction
The matrix metalloproteinase (MMP) family is a group
of zinc-binding endopeptidases that are involved in the breakdown
of extracellular matrix (1). The
majority of MMPs are secreted proteins that are activated by
extracellular proteinases. The membrane-type MMP (MT-MMP) subfamily
is a subtype of MMPs, which contain transmembrane domains and are
expressed on the surface of the cell membrane. MMP14 (also termed
MT1-MMP) is a member of this subfamily, containing a C-terminal
hydrophobic stretch to anchor the protein to the cell membrane
surface and to exert its effect. MMP14 activates MMP2, which is
closely associated with the invasion and metastasis of cancer
(2,3). MMP14 has also been demonstrated to be
associated with angiogenesis and cell migration (4,5).
Wickramasinghe et al (6)
reported the isolation of a peptide aptamer termed swiggle that
interacts with the intracellular domain of MMP14 and
swiggle-mediated inhibition of clathrin-mediated MMP14
internalisation was identified to promote MMP14-mediated cell
migration. MMP14 is overexpressed in numerous types of cancer and
is associated with a poor prognosis of pancreatic cancer (7,8),
hepatocellular carcinoma (9), lung
carcinomas (10–13), gastrointestinal carcinomas (14–17),
breast carcinomas (12,18,19),
gliomas (20) and cervical
carcinomas (21). We previously
analyzed the expression of MMP14 in pancreatic cancer tissues and
surrounding normal pancreatic tissues, which revealed that MMP14
was highly expressed in the pancreatic cancer group, while the
surrounding pancreatic tissue had a low expression level (22). Therefore, MMP14 has great application
prospects as a tumor biomarker (23). Previously, there have been a number
of reports of MMP14-specific functional blocking antibodies
(4,24–27) and
nanosensors (28), which indicate
MMP14 may be a useful tool for diagnostic and therapeutic
applications.
Aptamers can bind to target molecules with high
specificity via their specific three-dimensional structures;
therefore, aptamers are chemical antibodies (29). More specifically, nucleic acid
aptamers include short DNA and RNA aptamers. Cell-systematic
evolution of ligands by exponential enrichment (SELEX) is an
improvement of the conventional SELEX, which uses live cells as the
screening object to obtain aptamers of target molecules in the
natural state of the cell surface (29–32). In
order to reduce non-specific aptamers, negative screening cells are
usually introduced (29). Compared
with antibodies, aptamers have the advantages of cost, easy
synthesis, controllable modification, low toxicity, low
immunogenicity, long-term stability and a small size (33). Based on these advantages, aptamers
have become a popular topic in molecular imaging and targeted
therapy (34–40). For example, by modifying the
superparamagnetic iron oxide nanoparticle (SPION) and doxorubicin
co-loaded polymer with aptamer AS1411 as a targeting agent, the
probe could be used not only for magnetic resonance imaging (MRI)
of colon carcinoma xenografts, but also as a tumor-targeted
delivery system (36). Therefore,
aptamers exhibit great application prospects in molecular imaging
and targeted therapy of tumors as targeted tracers.
In the present study, 293T cells transfected with
the recombinant MMP14 gene were used as target cells to obtain the
DNA aptamer M17, which specifically recognizes MMP14-positive cells
through cell-SELEX technology, and its application potential in
imaging was further investigated. Aptamer M17 could be used to
target pancreatic cancer xenografts in fluorescent imaging. In
addition, M17 aptamer was conjugated to the surface of SPIONs
through a biotin-streptavidin system and the probe could
effectively reduce the T2-weighted MRI signal intensity of two
pancreatic cancer cell lines in vitro.
Materials and methods
Cell lines and cell culture
The 293T, PANC-1, MIA PaCa-2, HepG2 and MCF-7 cell
lines were provided by the Department of Biochemistry and Molecular
Biology, The Air Force Medical University (Xi'an, China). All cell
lines were grown in Dulbecco's modified Eagle's medium (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal
bovine serum (Thermo Fisher Scientific, Inc.) and incubated in 5%
CO2 at 37°C.
Construction of recombinant vector and
transfection
MMP14 complementary DNA was synthesized and spliced
into CD510B-1 plasmids (System Biosciences, LLC., Palo Alto,
California, USA). The sequence of the synthetic plasmid,
CD510B-1-MMP14, was confirmed through DNA sequencing by Sangon
Biotech Co., Ltd. (Shanghai, China). A total of 5 µg CD510B-1-MMP14
plasmids were transfected into 293T cells in a 60-mm dish using 15
µl TransEasy transfection reagent (Foregene, Shanghai, China).
Untreated CD510B-1 plasmids were transfected into 293T cells using
the same method and served as a control group termed 293T-Plasmid
cells. Following 36 h of transfection, the expression of MMP14 in
the transfected cells was confirmed by western blotting using an
anti-MMP14 antibody (Abcam, Cambridge, UK).
SELEX single-stranded DNA (ssDNA)
library and primers
The random sequence library and primers were
synthesized and purified by Sangon Biotech Co., Ltd. The random
sequence library consisted of three parts; the random part,
composed of 40 randomized nucleotides located in the center of the
sequence, and two constant parts, composed of 20 nucleotides
respectively located at both ends of the sequence. The sequence was
as follows: 5′-TGCGGAAGCCACCAGGAGTT(40N)ACGAGCCAAAGAGCCGCCAA-3′,
where 40N indicates the 40 randomized nucleotides. The forward
primer sequence was 5′-TGCGGAAGCCACCAGGAGT-3′ and its 5′ end was
labeled with 5-carboxyfluorescein (FAM). The reverse primer
sequence was 5′-TTGGCGGCTCTTTGGCTCGT-3′, which was labeled with
biotin at the 5′ end.
Cell-SELEX
The initial random sequence library of 5 nM was
dissolved in 200 µl binding buffer (Genshare Biological, Xi'an,
China) and then denatured for 5 min at 95°C, followed by immediate
cooling for 10 min on ice. Prior to screening, the 293T-MMP14 cells
were gently washed twice with PBS for 3 min. The cells
(~0.5×105/cm2) were incubated for 2 h with
the library at 37°C. To remove the unbound sequences, following
incubation the cells were gently washed three times for 3 min
washing buffer (Genshare Biological). The cells were collected and
kept in a 95°C water bath for 5 min and then immediately cooled for
10 min on ice. The cell-bound ssDNAs were extracted using phenol,
chloroform and isopropanol. The cell-bound ssDNA was precipitated
by sodium acetate and pre-cooled with isopropanol for 3 h at −20°C.
The sediment was dissolved in 200 µl ultrapure water. This solution
was then amplified by polymerase chain reaction (PCR) using Taq
polymerase and dNTPs from Takara Biotechnology Co., Ltd. (Dalian,
China). The PCR conditions were as follows: 95°C for 5 min,
followed by 30 cycles of 95°C for 10 sec, 57°C for 10 sec, 72°C for
10 sec and 72°C for 5 min. The PCR product was co-incubated with
streptavidin-labeled magnetic beads (BeaverBeads, Suzhou, China)
and the double-stranded DNA sequences were bound to the magnetic
beads via biotin-streptavidin. Following denaturation in 0.15 M
NaOH, the positive strand of DNA was dissociated, collected and
used for the next round of screening. The first round of screening
using the library labeled with FAM was obtained via asymmetric PCR
as aforementioned, except that the forward primers were labeled
with FAM, and flow cytometry, performed as aforementioned, was used
to monitor the effect of screening. From the third to the twelfth
round, the next-round library was incubated with the cells of the
control group and the unbound sequences were collected and
incubated with the target cells. In order to obtain
high-specificity and high-affinity aptamers, with increasing rounds
of screening, the incubation time with the target cells was
gradually decreased from 120 to 30 min, while the time with the
cells of the control group was gradually increased from 30 min to 2
h. The washing time following incubation with the target cells was
gradually increased. After 12 rounds of screening, the screened
ssDNA library was cloned into the plasmid using a M5 HiPer
pTOPO-Blunt Cloning kit (Mei5 Biotechnology, Co., Ltd, Beijing,
China). Subsequently, the products were transformed into DH5α
competent cell-coated plates for monoclonal colony selection and
sequencing by Sangon Biotech Co., Ltd. The secondary structure of
the finally obtained aptamers were predicted using the RNA
structure. To do so, first open the URL (http://rna.urmc.rochester.edu/RNAstructureWeb/),
then enter the aptamer name and sequence, select the nucleic acid
type as DNA, and finally click the ‘submit query’ to obtain the
predicted secondary structure of the aptamers.
Flow cytometry
After cells were detached by treatment with 0.1%
trypsin-0.02% EDTA solution, cells were placed in fetal calf serum
(FCS)-supplemented medium to inactivate trypsin and EDTA. Then the
cells were treated with labeling solution (Genshare Biological) at
37°C for 30 min. In total, ~2×105 cells were incubated
for 30 min at 37°C in the dark with the FAM-labeled random sequence
library or next round library (20 nM) or aptamers (2 nM) in PBS
binding buffer (200 µl) containing 10% FCS and 0.02% NaN3. After
incubation, the cells were washed thrice using 1,000 µl PBS washing
buffer containing 10% FCS, then re-suspended in 200 µl binding
buffer. The incubated cells were analyzed for FAM using a flow
cytometer. FlowJo7.0 software (version 7.0; FlowJo LLC, Ashland,
OR, USA) was used for the analysis of the flow cytometry data. The
dissociation constant (Kd) of the aptamer and target cells was
quantified using the one-site saturation equation: Y=Bmax × X/(Kd +
X), where Y is fluorescence intensity of the cells and X is
concentration of the aptamer and Bmax is maximum binding potential.
Analysis was performed used Sigma Plot 12.5 software (http://www.sigmaplot.co.uk/products/sigmaplot/sigmaplot-details.php).
Laser scanning confocal microscopy
(LSCM)
In total, ~1×105 293T-MMP14 cells or
293T-Plasmid cells were seeded in 35-mm dishes for LSCM. When the
cells reached 80% confluency, the cell culture medium was
discarded. Following three gentle washes with PBS, the cells were
fixed for 15 min with 4% paraformaldehyde at room temperature, and
then blocked for 30 min with 1% bovine serum albumin at room
temperature. The cells were then incubated for 30 min with 20 nM
FAM-labeled aptamers at 37°C and the cell nucleus was stained with
DAPI for 5 min at room temperature. The cells were finally imaged
by LSCM (magnification, ×800). Pancreatic cancer cells were seeded
and cultured in the same way. Following washing with PBS, the
target cells were incubated for 30 min with 20 nM Cy5-labeled
aptamers at 37°C and then images were obtained by LSCM
(magnification ×400).
Western blot analysis
The western blot analysis was performed in
accordance with the protocol by Abcam. Proteins were extracted
using 10× RIPA Buffer (Abcam) and determined using the BCA method.
Proteins were loaded onto a polyacrylamide gel (10% gel) at a mass
of 20 µg per lane, and separated via constant pressure
electrophoresis. The concentration of the gel was 10%. The protein
in the gel was transferred under constant voltage to a
polyvinylidene difluoride (PVDF) membrane. Following blocking for 1
h at room temperature with PBS containing 5% skimed milk, the PVDF
membrane was incubated with anti-MMP14 antibody (1:5,000; ab51074;
Abcam) at 4°C overnight. After washing three times with TBS
containing 0.1% tween, the membrane was incubated with the
secondary antibody labeled with horseradish peroxidase (HRP;
1:10,000; ab6721; Abcam) for 1 h at room temperature. Finally, the
immunoreactive bands were visualized by a Chemiluminescent HRP
Substrate (Abcam).
In vivo fluorescence imaging
A total of 30 Athymic BALB/c, 4-week-old mice (15
male, 15 female) were provided by the Experimental Animal Center of
the Air Force Medical University (Xi'an, China). The average weight
of the males was 17 g, and the average weight of the females was 14
g. The housing criteria of the mice was set up as per guidelines
from the Experimental Animal Center of the Air Force Medical
University. The tumor-bearing nude mouse model was generated by
subcutaneously injecting 1×107 MIA PaCa-2 cells in
suspension into the axilla of the mice. Tumors were observed until
they reached 1.0 cm in diameter. The mice were anesthetized using a
small animal anesthesia system (E-Z Anesthesia EZ-7000 classic
system, E-Z Anesthesia, Palmer, PA, USA). The animal study was
approved by the Ethics Committee of the Experimental Animal Center
of the Air Force Medical University. When the mice were
anesthetized, 100 nM Cy5-labeled aptamer M17 or the initial library
was injected through the tail vein. At different time points, the
Cy5 fluorescence signal of the mice was acquired by a whole body
imaging system (IVIS Lumina II Series, Caliper Life Sciences;
PerkinElmer, Inc., Waltham, MA, USA).
Synthesis of aptamer-conjugated
magnetic nanoparticles and in vitro MRI
Streptavidin-coated polyethylene glycol (PEG)-Fe3O4
nanoparticles (Nanjing Nanoeast Biological Technology Co., Ltd.,
Nanjing, China) were reacted with biotin-labeled M17 and the
unselected initial library separately for 1 h on a shaker to
generate the M17-SPIONs and Lib-SPIONs, respectively, through a
biotin-streptavidin system. The probes were gathered by magnetic
separation and finally re-suspended in 3% agarose gel to the
required concentration. The relaxivity of the magnetic
nanoparticles was measured using a Siemens 3.0T MRI scanner
(Siemens AG, Munich, Germany) when the solution was solidified. The
T2 weighted imaging (T2WI) measurement parameters were as follows:
Time of repetition, 3,500 ms; time of echo, 91 ms; averages, 8; and
field of field, 100 mm. TR is time of repetition, TE is time of
echo, FOV is field of view. Subsequently, ~1×107 target
cells were incubated with M17-SPIONs and Lib-SPIONs at 37°C for 1
h. Then the cells were washed thrice with PBS and resuspended in 3%
agarose gel and scanned with an MRI scanner when the solution was
solidified. 293T cells were used as a negative control.
Statistical analysis
All measurement data is presented as the mean ±
standard deviation. The data were analyzed using a two-sample
Student's t-test and Pearson's correlation analysis using SPSS
software (version 17.0; SPSS, Inc., Chicago, IL, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results and Discussion
Expression of MMP14 in transfected
cells
To effectively screen aptamers that specifically
recognize MMP14, MMP14 was overexpressed in 293T cells while
maintaining its native conformation on the cell membrane. The cells
of the negative control group expressed a low level of MMP14.
293T-MMP14 cells and 293T-Plasmid cells were obtained by
transfecting CD510B-1-MMP14 and CD510B-1 plasmids, respectively,
into 293T cells. A total of 36 h after transfection, western
blotting revealed that MMP14 was overexpressed in 293T-MMP14 cells
and expressed at an almost undetectable level in 293T-Plasmid cells
(Fig. 1). This result indicated that
two transfected cell lines were generated and there was a marked
difference in the expression of MMP14 between them.
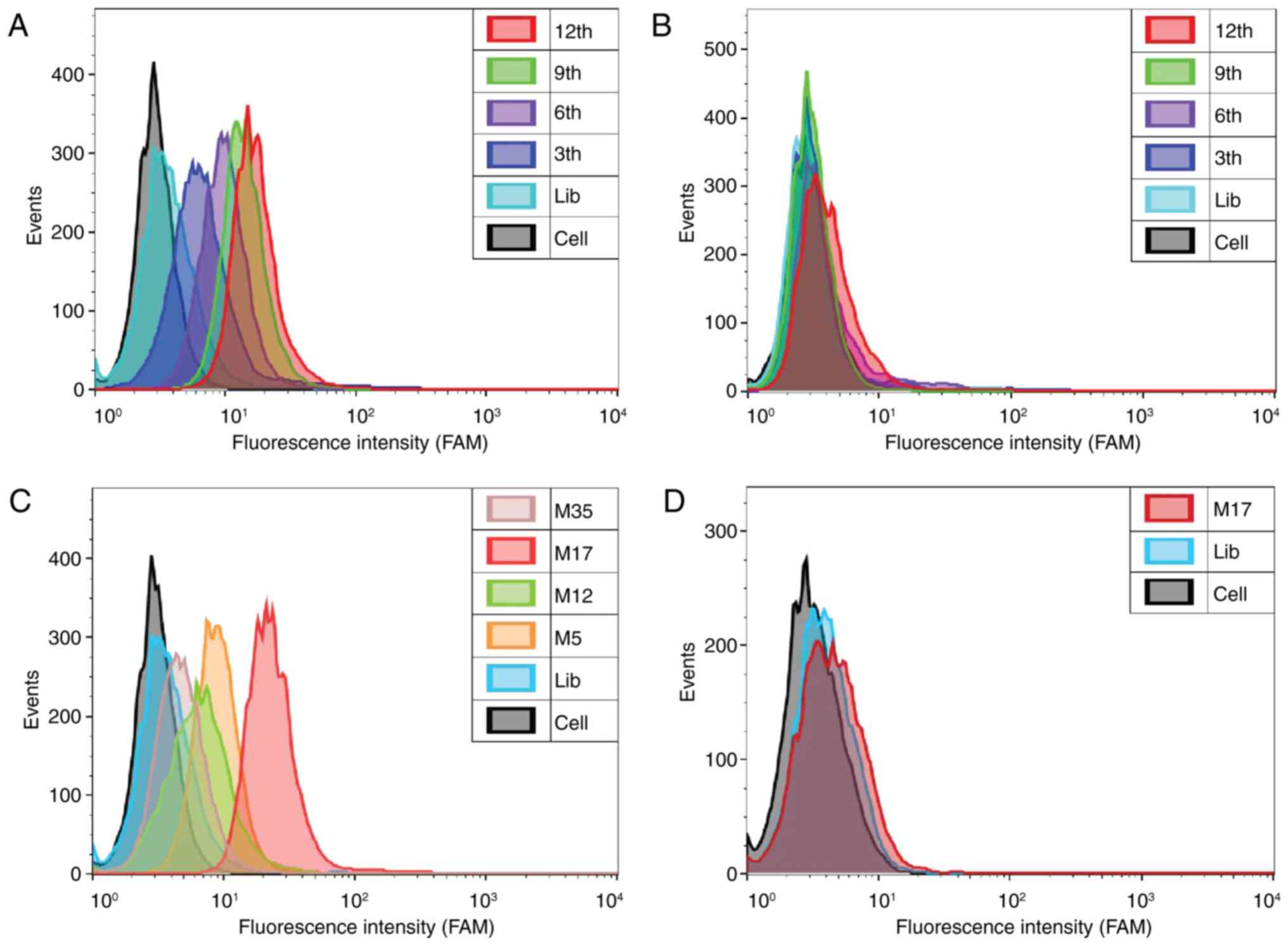
Screening results of DNA aptamers
In the cell-SELEX, 293T-MMP14 cells were used as
positive screening cells. To reduce non-specific sequences,
293T-Plasmid cells were used as negative control cells. The
enrichment effect of each selected round was monitored by flow
cytometry. The fluorescence intensity of the cells indicated the
binding affinity of the library. The fluorescence intensity of the
293T-MMP14 cells bound to the selected library increased with the
number of selection rounds. However, no increase was observed in
the 293T-Plasmid cells (Fig. 2A and
B). The percentages of fluorescent-labeled cells at various
stages of the process are presented in Table I. The results indicated that the
ssDNA sequences that specifically bound to the target cells were
gradually enriched. However, the degree of increase gradually
decreased as the number of selection rounds increased. The
screening process was completed after 12 rounds. The final library
was amplified and cloned, then sequenced by Tsingke Biological
Technology Co., Ltd. (Beijing, China).
 | Table I.Percentage of fluorescent-labeled
cells for cell-systematic evolution of ligands by exponential
enrichment. |
Table I.
Percentage of fluorescent-labeled
cells for cell-systematic evolution of ligands by exponential
enrichment.
|
| Ratio of positive
cells, % |
|---|
|
|
|
|---|
|
| 293T-MMP14 | 293T-Plasmid |
|---|
| Cell |
0.16 | 1.38 |
| Lib |
3.65 | 4.80 |
| 3th | 19.5 | 6.24 |
| 6th | 48.7 | 8.66 |
| 9th | 84.5 | 1.57 |
| 12th | 95.3 | 1.38 |
Analysis and verification of ssDNA
aptamers
After sequencing, 24 effective sequences were
selected and synthesized by Sangon Biotech Co., Ltd. with a FAM
label on the 5′ end. Four representative sequences (M5, M12, M17
and M35) were selected by flow cytometry due to their high affinity
for recognizing 293T-MMP14 cells. Among these, aptamer M17
demonstrated the highest affinity for 293T-MMP14 cells compared
with 293T-Plasmid cells (Fig. 2C and
D). When cells were incubated with the FAM-labeled aptamer M17,
the confocal imaging results demonstrated that the fluorescence
signal was predominantly located around the 293T-MMP14 cells and
not the 293T-Plasmid cells (Fig. 3).
These results indicated that aptamer M17 discriminated 293T-MMP14
cells from control cells and that the binding site of aptamer M17
was located on the cell membrane. The Kd value of M17 to 293T-MMP14
cells was 4.98±1.26 nM (Fig. 4A),
indicating that M17 could recognize 293T-MMP14 cells with high
specificity and high affinity. The secondary structure of aptamer
M17 (Fig. 4B) was predicted using
RNAstructure (http://rna.urmc.rochester.edu/RNAstructureWeb/). The
sequence of aptamer M17 is as follows:
5′-AGGGCCCGACGTGACGGCACGTCGGATATCTCATGCGTGT-3′.
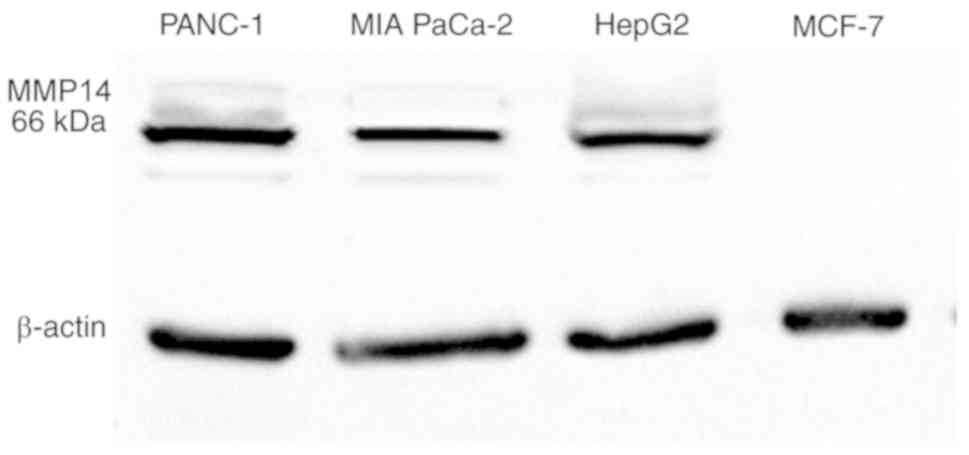
To investigate the binding ability of aptamer M17
for MMP14-positive cell lines, PANC-1, MIA PaCa-2, HepG2 and MCF-7
were incubated with aptamer M17 labeled with FAM. The initial
library labeled by FAM was used as a negative control. Western
blotting revealed that PANC-1, MIA PaCa-2 and HepG2 cells highly
expressed MMP14, while MCF-7 cells expressed an almost undetectable
level of MMP14 (Fig. 5). As
presented in Fig. 6, aptamer M17
markedly increased the fluorescence signal in pancreatic cancer
(PANC-1, MIA PaCa-2) cells and hepatoblastoma (HepG2) cells, but
not in breast cancer (MCF-7) cells. The selected initial library
did not have this effect. These results indicate that the aptamer
M17 could distinguish the MMP14-positive cancer cell lines from the
MMP14-negative cell lines. However, it is not clear which part of
MMP14 interacts with aptamer M17. Therefore, this was the main area
of focus in the rest of the present study.
As presented in Fig.
7, The MIA PaCa-2 cells and PANC-1 cells were incubated with
M17 labeled with Cy5 at 37°C for 30 min. The results of confocal
microscopy imaging revealed that M17 labeled by Cy5 specifically
targeted the surface of the pancreatic cancer cell membrane.
In vivo fluorescence imaging for
pancreatic tumor-bearing mice
To verify that the aptamer M17 had the ability to
recognize MMP14 with high specificity in vivo, MIA
PaCa-2-cell tumor-bearing nude mice were used. After the
Cy5-labeled aptamer M17 was intravenously injected into the
tumor-bearing nude mice for 3 min, a fluorescence signal was
observed at the tumor site, which disappeared after 40 min. The
Cy5-labeled initial library was also injected into the
tumor-bearing nude mice, after which no fluorescence signal was
evident at the tumor site at any observation time point (Fig. 8). These results indicate that the
aptamer M17 could specifically recognize MIA PaCa-2 cells
(MMP14-positive) in vivo, indicating that the aptamer M17
has a potential application for recognizing MMP14-positive cancer
in vivo. The targeting residence time of aptamer M17 in the
tumor-bearing nude mice was limited and completely disappeared
after 40 min. The reason for this may be that the aptamer M17 was
an unmodified aptamer. However, aptamer M17 successfully achieved
targeted imaging of tumors. The next step may be to modify the
aptamer to improve its nuclease resistance.
Synthesis of aptamer M17-conjugated
magnetic nanoparticles and in vitro MRI
To verify the feasibility of SPIONs as MRI T2WI
contrast agents, T2WI using different concentrations of SPIONs was
performed using a 3.0T MRI scanner and the T2WI values were
measured through the workstation. Fig.
9C presents T2WI images of a 3% agarose gel model with
different concentrations of SPIONs. As the concentration of SPIONs
increased, the brightness of the T2WI images decreased. Fig. 9A presents the correlation between
SPIONs concentration and the T2WI value. The correlation
coefficient (r=−0.738) indicated that the concentration of SPIONs
was significantly negatively correlated with the T2WI value. This
result indicates that SPIONs were effective T2WI darkening contrast
agents.
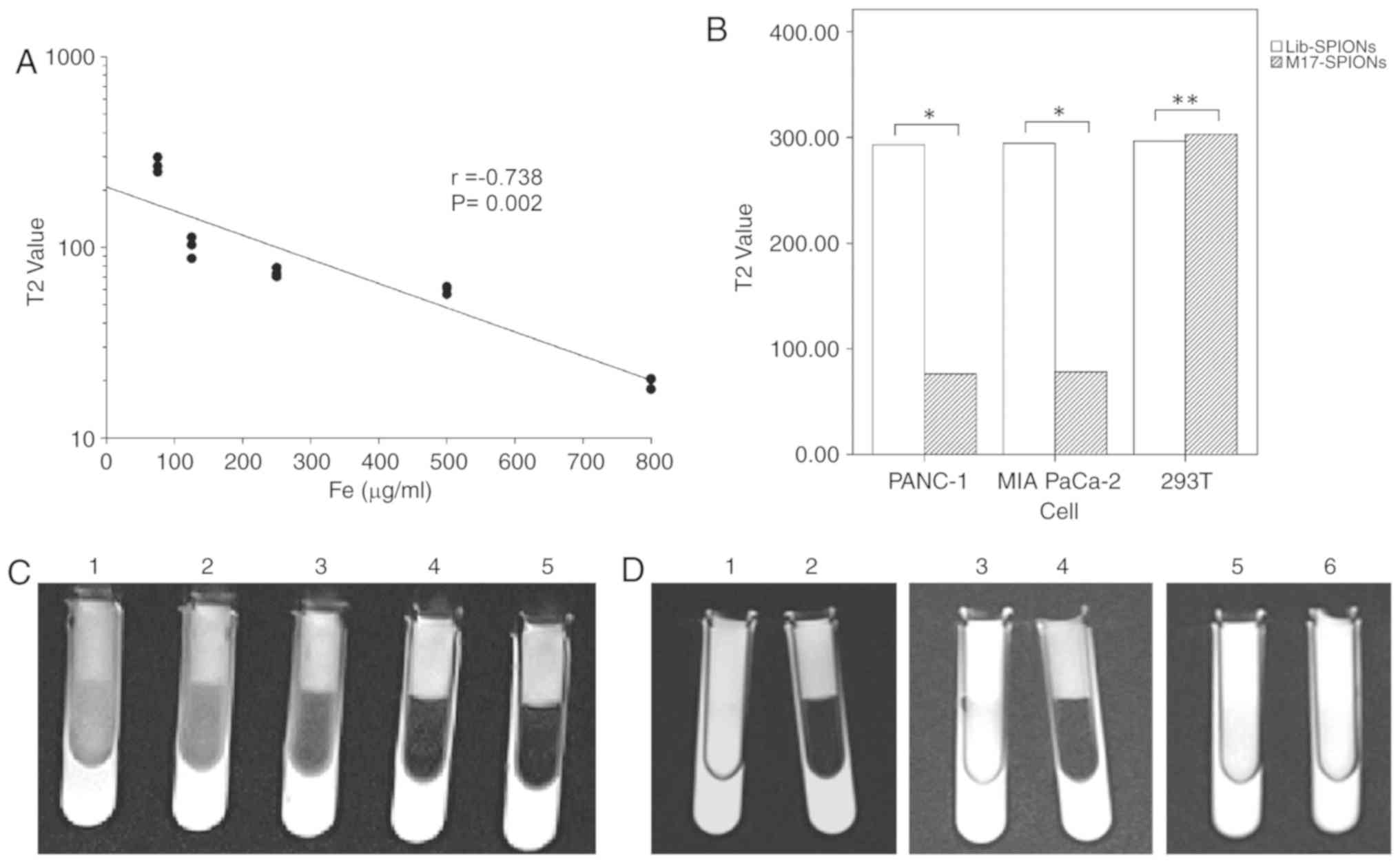 | Figure 9.MRI measurements. (A) Correlation
between T2 value and SPIONs concentration. The coefficient was
r=−0.738 (P=0.002), as determined by Pearson's correlation
analysis. (B) The T2 values of different cell groups incubated with
M17-SPIONs or Lib-SPIONs. (C) T2WI images of different
concentrations of SPIONs scanned by 3.0T MRI. The concentrations of
SPIONs added to centrifugal tubes nos. 1–5 were 60, 120, 250, 500
and 800 µg/ml, respectively. (D) T2WI images of different cell
lines incubated with M17-SPIONs or Lib-SPIONs scanned by 3.0T MRI.
Centrifugal tubes 1 and 2 contained 293T cells, 3 and 4 contained
Panc-1 cells, and 5 and 6 contained MIAPaCa-2 cells. Centrifugal
tubes 1, 3 and 5 were incubated with SPIONs-M17, and 2, 4 and 6
were incubated with SPIONs-Lib. *P<0.01, **P>0.01. MRI,
magnetic resonance imaging; SPION, superparamagnetic iron oxide
nanoparticle; Lib, initial library; Fe, iron. |
To demonstrate the ability of M17-conjugated SPIONs
(M17-SPIONs) to target pancreatic cancer cells for MRI in
vitro, cells were incubated with M17-SPIONs probes at 37°C for
1 h. Subsequently, the cells were resuspended in 3% agarose gel.
The T2WI images of different cells were scanned using a 3.0T MRI
scanner. The Lib-SPIONs probes served as a negative control.
Fig. 9D presents T2WI images of the
different cell groups. The T2WI images of MIA PaCa-2 cells and
PANC-1 cells incubated with M17-SPIONs were markedly darkened,
while those of 293T cells were not. After incubation with
Lib-SPIONs, the T2WI images did not darken for any cell group.
Fig. 9B presents a histogram of T2WI
values of the different cell groups incubated with two probes.
These results indicate that M17-SPIONs effectively reduced T2WI
values in MIA PaCa-2 and PANC-1 cells in vitro. The
M17-SPIONs probe was demonstrated to be a potential MRI nanoprobe
for pancreatic cancer. In the future, the conditions of probes
(M17- SPIONs) applied in in vivo xenograft models should be
optimized.
In conclusion, aptamer M17, which specifically
recognized MMP14-positive cancer cells, was successfully obtained
by Cell-SELEX after 12 rounds of screening. Aptamer M17 could bind
to MMP14-transfected 293T cells with high specificity and high
affinity, with a Kd value in the nanomolar range. Binding analysis
revealed that aptamer M17 can recognize MMP14-positive cancer
cells, including PANC-1, MIA PaCa-2 and HepG2 cells. Tumor imaging
in vivo demonstrated that aptamer M17 has potential for
targeted diagnosis and treatment of pancreatic cancer. In addition,
aptamer M17-conjugated SPIONs (M17-SPIONs) demonstrated efficient
targeted MRI of pancreatic cancer cells in vitro. In
summary, DNA aptamer M17 is a promising molecular targeting agent
and has potential application value in the targeted diagnosis and
treatment of MMP14-positive cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. NSFC 81220108011
and NSFC 81370039).
Availability of data and materials
Not applicable.
Authors' contributions
XH designed the experiment, wrote the manuscript,
performed aptamer screening and managed the team. JZ cultured the
cells, monitored the mice and test cells, and performed flow
cytometry and fluorescence imaging. JR analyzed the data and
reviewed the manuscript. DW constructed the tumor-bearing nude mice
model and performed in vivo imaging of the mice. WZ
performed magnetic resonance imaging of the cells. YH was
responsible for assessing the feasibility of the study design,
guiding the experimental process and the assessment of data
accuracy. In addition, YH agreed to be accountable for all aspects
of the work in ensuring that questions related to the accuracy or
integrity of any part of the work are appropriately investigated
and resolved and gave final approval of the manuscript version to
be published. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
the Experimental Animal Center of the Fourth Military Medical
University (Xi'an, China). The relevant proofs are filed at the
Animal Experimental Center.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Brinckerhoff CE and Matrisian LM: Matrix
metalloproteinases: A tail of a frog that became a prince. Nat Rev
Mol Cell Biol. 3:207–214. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Suzuki A, Lu J, Kusakai G, Kishimoto A,
Ogura T and Esumi H: ARK5 is a tumor invasion-associated factor
downstream of Akt signaling. Mol Cell Biol. 24:3526–3535. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Seiki M: Membrane-type 1 matrix
metalloproteinase: A key enzyme for tumor invasion. Cancer Lett.
194:1–11. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Galvez BG, Matias-Roman S, Albar JP,
Sanchez-Madrid F and Arroyo AG: Membrane type 1-matrix
metalloproteinase is activated during migration of human
endothelial cells and modulates endothelial motility and matrix
remodeling. J Biol Chem. 276:37491–37500. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arroyo AG, Genis L, Gonzalo P,
Matias-Roman S, Pollan A and Galvez BG: Matrix metalloproteinases:
New routes to the use of MT1-MMP as a therapeutic target in
angiogenesis-related disease. Curr Pharm Des. 13:1787–1802. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wickramasinghe RD, Ko Ferrigno P and Roghi
C: Peptide aptamers as new tools to modulate clathrin-mediated
internalisation-inhibition of MT1-MMP internalisation. BMC Cell
Biol. 11:582010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ottaviano AJ, Sun L, Ananthanarayanan V
and Munshi HG: Extracellular matrix-mediated membrane-type 1 matrix
metalloproteinase expression in pancreatic ductal cells is
regulated by transforming growth factor-beta1. Cancer Res.
66:7032–7040. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Imamura T, Ohshio G, Mise M, Harada T,
Suwa H, Okada N, Wang Z, Yoshitomi S, Tanaka T, Sato H, et al:
Expression of membrane-type matrix metalloproteinase-1 in human
pancreatic adenocarcinomas. J Cancer Res Clin Oncol. 124:65–72.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen TY, Li YC, Liu YF, Tsai CM, Hsieh YH,
Lin CW, Yang SF and Weng CJ: Role of MMP14 gene polymorphisms in
susceptibility and pathological development to hepatocellular
carcinoma. Ann Surg Oncol. 18:2348–2356. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sato H, Takino T, Okada Y, Cao J,
Shinagawa A, Yamamoto E and Seiki M: A matrix metalloproteinase
expressed on the surface of invasive tumour cells. Nature.
370:61–65. 1994. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tokuraku M, Sato H, Murakami S, Okada Y,
Watanabe Y and Seiki M: Activation of the precursor of gelatinase
A/72 kDa type IV collagenase/MMP-2 in lung carcinomas correlates
with the expression of membrane-type matrix metalloproteinase
(MT-MMP) and with lymph node metastasis. Int J Cancer. 64:355–359.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Polette M, Nawrocki B, Gilles C, Sato H,
Seiki M, Tournier JM and Birembaut P: MT-MMP expression and
localisation in human lung and breast cancers. Virchows Arch.
428:29–35. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nawrocki B, Polette M, Marchand V, Monteau
M, Gillery P, Tournier JM and Birembaut P: Expression of matrix
metalloproteinases and their inhibitors in human bronchopulmonary
carcinomas: Quantificative and morphological analyses. Int J
Cancer. 72:556–564. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mori M, Mimori K, Shiraishi T, Fujie T,
Baba K, Kusumoto H, Haraguchi M, Ueo H and Akiyoshi T: Analysis of
MT1-MMP and MMP2 expression in human gastric cancers. Int J Cancer.
74:316–321. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nomura H, Sato H, Seiki M, Mai M and Okada
Y: Expression of membrane-type matrix metalloproteinase in human
gastric carcinomas. Cancer Res. 55:3263–3266. 1995.PubMed/NCBI
|
|
16
|
Bando E, Yonemura Y, Endou Y, Sasaki T,
Taniguchi K, Fujita H, Fushida S, Fujimura T, Nishimura G, Miwa K
and Seiki M: Immunohistochemical study of MT-MMP tissue status in
gastric carcinoma and correlation with survival analyzed by
univariate and multivariate analysis. Oncol Rep. 5:1483–1488.
1998.PubMed/NCBI
|
|
17
|
Ohtani H, Motohashi H, Sato H, Seiki M and
Nagura H: Dual over-expression pattern of membrane-type
metalloproteinase-1 in cancer and stromal cells in human
gastrointestinal carcinoma revealed by in situ hybridization and
immunoelectron microscopy. Int J Cancer. 68:565–570. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ueno H, Nakamura H, Inoue M, Imai K,
Noguchi M, Sato H, Seiki M and Okada Y: Expression and tissue
localization of membrane-types 1, 2, and 3 matrix
metalloproteinases in human invasive breast carcinomas. Cancer Res.
57:2055–2060. 1997.PubMed/NCBI
|
|
19
|
Jones JL, Glynn P and Walker RA:
Expression of MMP-2 and MMP-9, their inhibitors, and the activator
MT1-MMP in primary breast carcinomas. J Pathol. 189:161–168. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang L, Yuan J, Tu Y, Mao X, He S, Fu G,
Zong J and Zhang Y: Co-expression of MMP-14 and MMP-19 predicts
poor survival in human glioma. Clin Transl Oncol. 15:139–145. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gilles C, Polette M, Piette J, Munaut C,
Thompson EW, Birembaut P and Foidart JM: High level of MT-MMP
expression is associated with invasiveness of cervical cancer
cells. Int J Cancer. 65:209–213. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang ZH, Wen DD, Fu X, Zhong JM, Lu JT,
Huang XF, Ren J, Yang Y and Huan Y: Study on the expression and
clinical significance of survivin and MMP14 in pancreatic cancer.
Progress in Modern Biomed. 15:3022–3027. 2015.(In Chinese).
|
|
23
|
Shimizu Y, Temma T, Sano K, Ono M and Saji
H: Development of membrane type-1 matrix metalloproteinase-specific
activatable fluorescent probe for malignant tumor detection. Cancer
Sci. 102:1897–1903. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Devy L, Huang L, Naa L, Yanamandra N,
Pieters H, Frans N, Chang E, Tao Q, Vanhove M, Lejeune A, et al:
Selective inhibition of matrix metalloproteinase-14 blocks tumor
growth, invasion, and angiogenesis. Cancer Res. 69:1517–1526. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ingvarsen S, Porse A, Erpicum C, Maertens
L, Jurgensen HJ, Madsen DH, Melander MC, Gardsvoll H, Hoyer-Hansen
G, Noel A, et al: Targeting a single function of the
multifunctional matrix metalloprotease MT1-MMP: Impact on
lymphangiogenesis. J Biol Chem. 288:10195–10204. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Haage A, Nam DH, Ge X and Schneider IC:
Matrix metalloproteinase-14 is a mechanically regulated activator
of secreted MMPs and invasion. Biochem Biophys Res Commun.
450:213–218. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Udi Y, Grossman M, Solomonov I, Dym O,
Rozenberg H, Moreno V, Cuniasse P, Dive V, Arroyo AG and Sagi I:
Inhibition mechanism of membrane metalloprotease by an
exosite-swiveling conformational antibody. Structure. 23:104–115.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chung E, Ochs CJ, Wang Y, Lei L, Qin Q,
Smith AM, Alex S, Kamm RD, Qi YX, Lu S and Wang YP: Activatable and
Cell-Penetrable Multiplex FRET Nanosensor for Profiling MT1-MMP
Activity in Single Cancer Cells. Nano Lett. 15:5025–5032. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sefah K, Shangguan D, Xiong X, O'Donoghue
MB and Tan W: Development of DNA aptamers using Cell-SELEX. Nat
Protoc. 5:1169–1185. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ellington AD and Szostak JW: In
vitro selection of RNA molecules that bind specific ligands.
Nature. 346:818–822. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tuerk C and Gold L: Systematic evolution
of ligands by exponential enrichment: RNA ligands to bacteriophage
T4 DNA polymerase. Science. 249:505–510. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shangguan D, Li Y, Tang Z, Cao ZC, Chen
HW, Mallikaratchy P, Sefah K, Yang CJ and Tan W: Aptamers evolved
from live cells as effective molecular probes for cancer study.
Proc Natl Acad Sci USA. 103:11838–11843. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu J, You M, Pu Y, Liu H, Ye M and Tan W:
Recent developments in protein and cell-targeted aptamer selection
and applications. Curr Med Chem. 18:4117–4125. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xing H, Wong NY, Xiang Y and Lu Y: DNA
aptamer functionalized nanomaterials for intracellular analysis,
cancer cell imaging and drug delivery. Curr Opin Chem Biol.
16:429–435. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hwang DW, Ko HY, Lee JH, Kang H, Ryu SH,
Song IC, Lee DS and Kim S: A nucleolin-targeted multimodal
nanoparticle imaging probe for tracking cancer cells using an
aptamer. J Nucl Med. 51:98–105. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mosafer J, Abnous K, Tafaghodi M,
Mokhtarzadeh A and Ramezani M: In vitro and in vivo evaluation of
anti-nucleolin-targeted magnetic PLGA nanoparticles loaded with
doxorubicin as a theranostic agent for enhanced targeted cancer
imaging and therapy. Eur J Pharm Biopharm. 113:60–74. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
You XG, Tu R, Peng ML, Bai YJ, Tan M, Li
HJ, Guan J and Wen LJ: Molecular magnetic resonance probe targeting
VEGF165: Preparation and in vitro and in vivo evaluation. Contrast
Media Mol Imaging. 9:349–354. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pu Y, Liu Z, Lu Y, Yuan P, Liu J, Yu B,
Wang G, Yang CJ, Liu H and Tan W: Using DNA aptamer probe for
immunostaining of cancer frozen tissues. Anal Chem. 87:1919–1924.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pilapong C, Sitthichai S, Thongtem S and
Thongtem T: Smart magnetic nanoparticle-aptamer probe for targeted
imaging and treatment of hepatocellular carcinoma. Int J Pharm.
473:469–474. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li CH, Kuo TR, Su HJ, Lai WY, Yang PC,
Chen JS, Wang DY, Wu YC and Chen CC: Fluorescence-guided probes of
aptamer-targeted gold nanoparticles with computed tomography
imaging accesses for in vivo tumor resection. Sci Rep.
5:156752015. View Article : Google Scholar : PubMed/NCBI
|