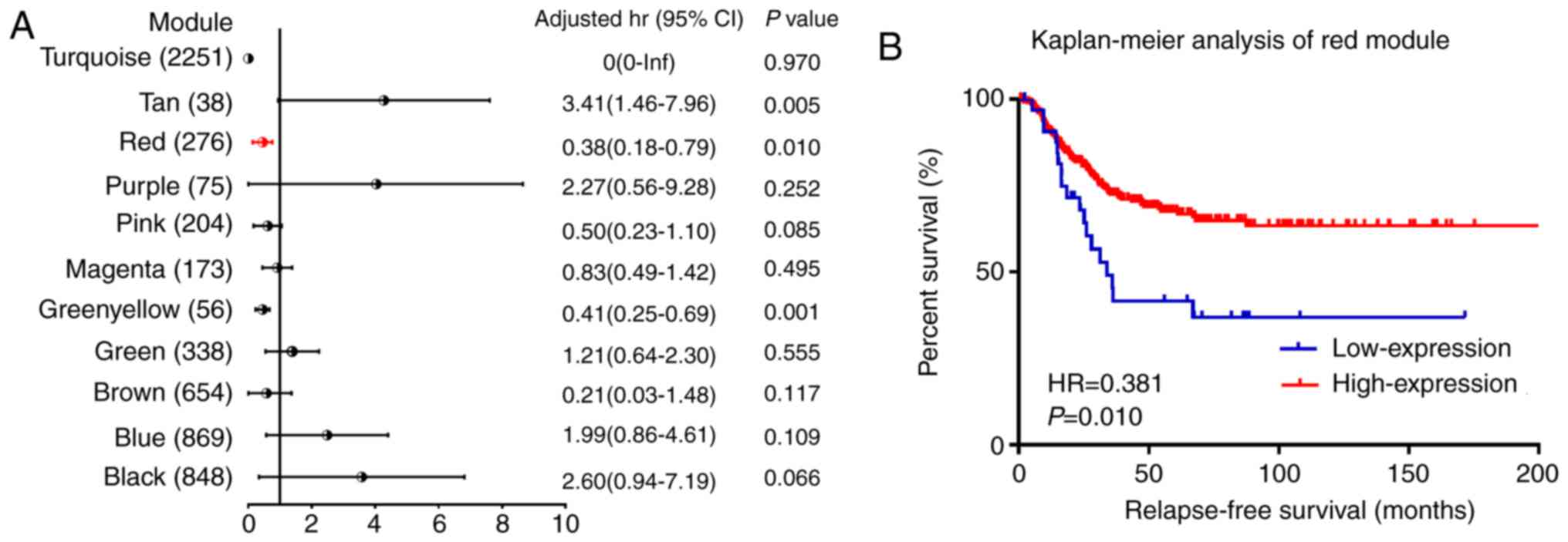
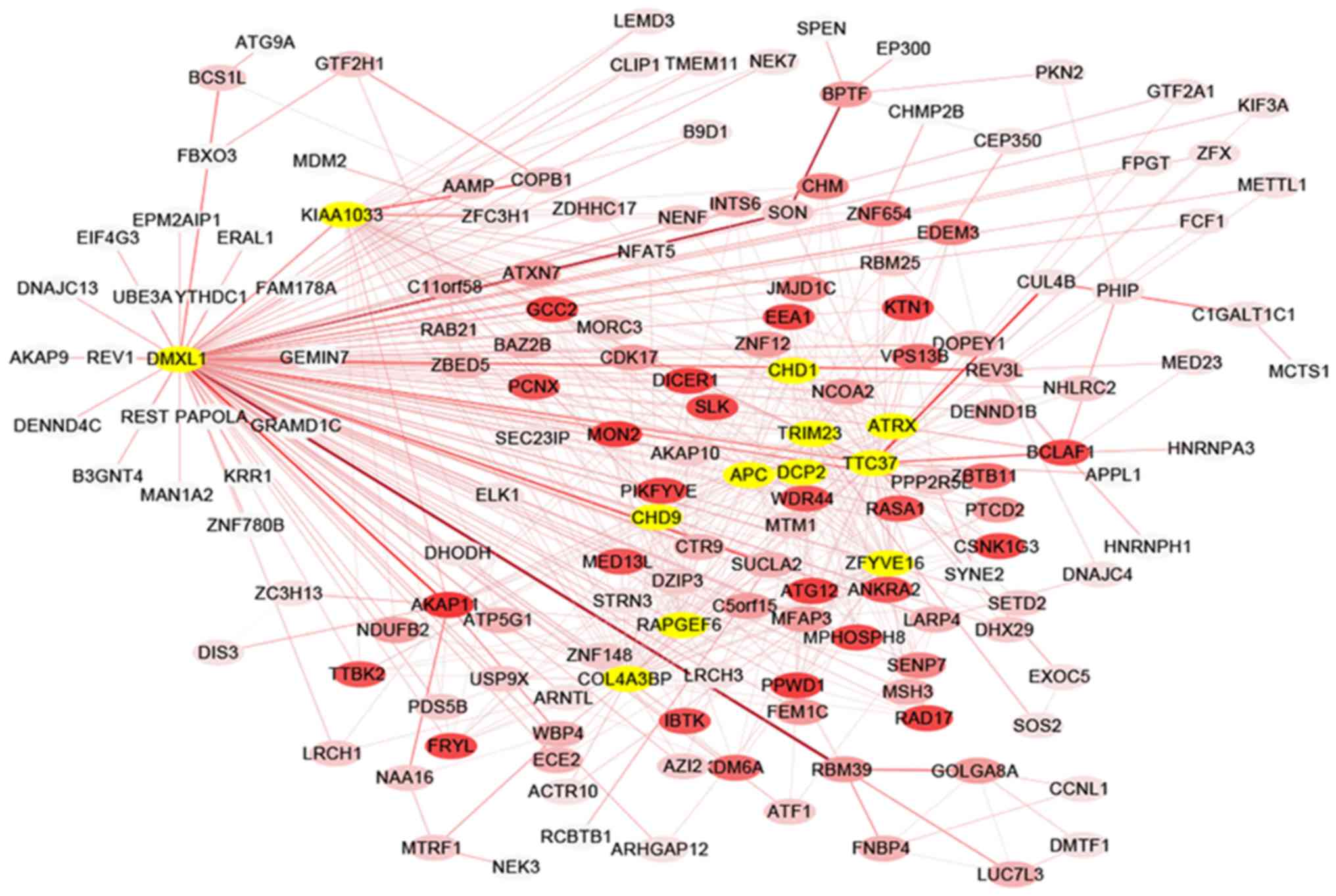
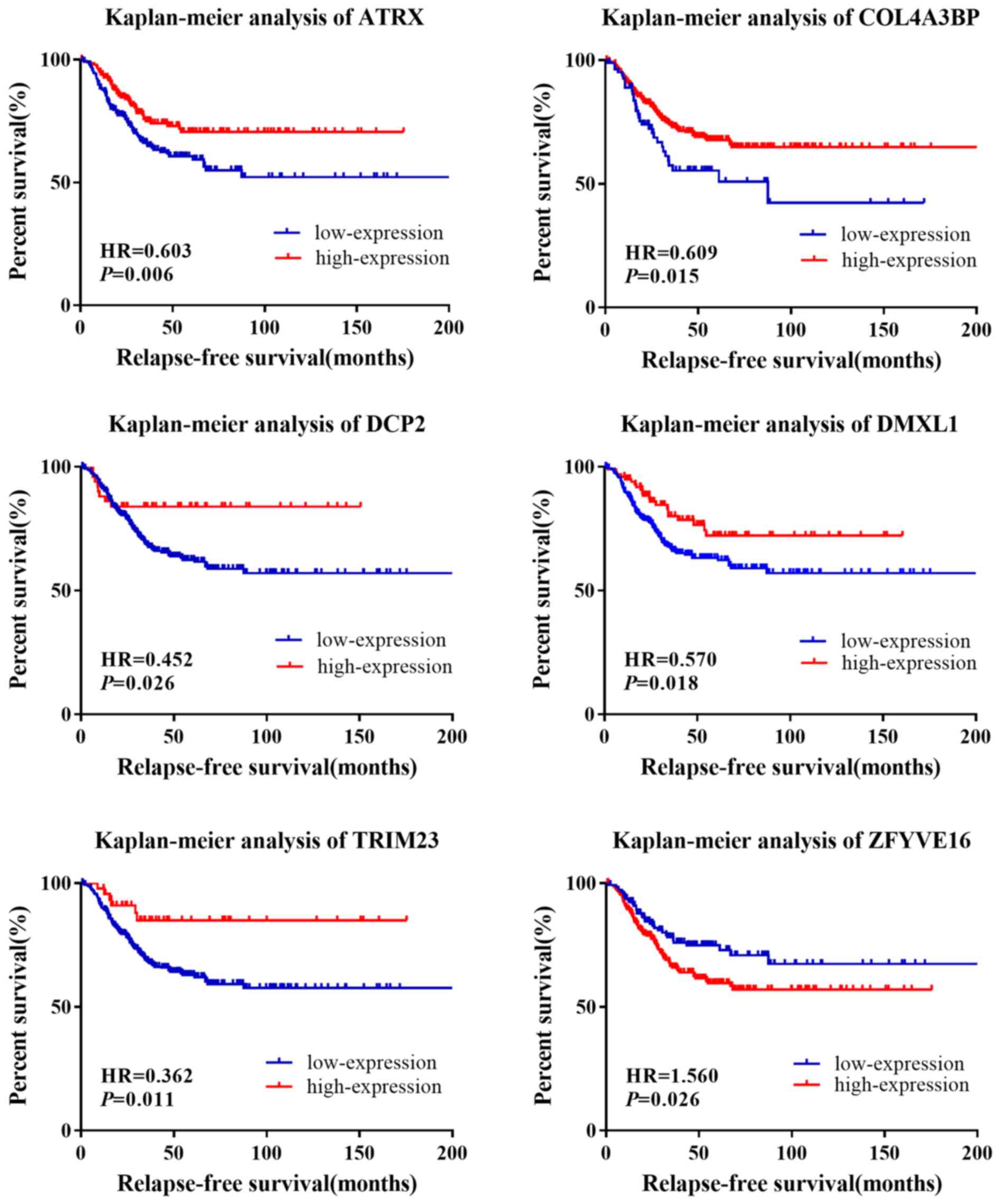
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kumar P and Aggarwal R: An overview of
triple-negative breast cancer. Arch Gynecol Obstet. 293:247–269.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aysola K, Desai A, Welch C, Xu J, Qin Y,
Reddy V, Matthews R, Owens C, Okoli J, Beech DJ, et al: Triple
negative breast cancer-an overview. Hereditary Genet 2013. (Suppl
2):0012013.
|
|
5
|
Saha P and Nanda R: Concepts and targets
in triple-negative breast cancer: Recent results and clinical
implications. Ther Adv Med Oncol. 8:351–359. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu YL, Yao R, Li J, Zhou YD, Mao F, Pan B
and Sun Q: FOXC1 overexpression is a marker of poor response to
anthracycline-based adjuvant chemotherapy in sporadic
triple-negative breast cancer. Cancer Chemother Pharmacol.
79:1205–1213. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Horvath S, Zhang B, Carlson M, Lu KV, Zhu
S, Felciano RM, Laurance MF, Zhao W, Qi S, Chen Z, et al: Analysis
of oncogenic signaling networks in glioblastoma identifies ASPM as
a novel molecular target. Proc Natl Acad Sci USA. 103:17402–17407.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang L, Tang H, Thayanithy V, Subramanian
S, Oberg AL, Cunningham JM, Cerhan JR, Steer CJ and Thibodeau SN:
Gene networks and microRNAs implicated in aggressive prostate
cancer. Cancer Res. 69:9490–9497. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang B and Horvath S: A general framework
for weighted gene co-expression network analysis. Stat Appl Genet
Mol Biol. 4:Article172005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao W, Langfelder P, Fuller T, Dong J, Li
A and Hovarth S: Weighted gene coexpression network analysis: State
of the art. J Biopharm Stat. 20:281–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen L, Yuan L, Wang Y, Wang G, Zhu Y, Cao
R, Qian G, Xie C, Liu X, Xiao Y and Wang X: Co-expression network
analysis identified FCER1G in association with progression and
prognosis in human clear cell renal cell carcinoma. Int J Biol Sci.
13:1361–1372. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wan Q, Tang J, Han Y and Wang D:
Co-expression modules construction by WGCNA and identify potential
prognostic markers of uveal melanoma. Exp Eye Res. 166:13–20. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Clarke C, Madden SF, Doolan P, Aherne ST,
Joyce H, O'Driscoll L, Gallagher WM, Hennessy BT, Moriarty M, Crown
J, et al: Correlating transcriptional networks to breast cancer
survival: A large-scale coexpression analysis. Carcinogenesis.
34:2300–2308. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Langfelder P and Horvath S: WGNCA: an R
package for weighted correlation network analysis. BMC Genetics.
9:5592008.
|
|
16
|
Chen F, Zhu HH, Zhou LF, Li J, Zhao LY, Wu
SS, Wang J, Liu W and Chen Z: Genes related to the very early stage
of ConA-induced fulminant hepatitis: A gene-chip-based study in a
mouse model. BMC Genomics. 11:2402010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu X, Hu AX, Zhao JL and Chen FL:
Identification of key gene modules for in human osteosarcoma by
co-expression analysis weighted gene co-expression network analysis
(WGCNA). J Cell Biochem. 118:3953–3959. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yuan L, Chen L, Qian K, Wang G, Lu M, Qian
G, Cao X, Jiang W, Xiao Y and Wang X: A novel correlation between
ATP5A1 gene expression and progression of human clear cell renal
cell carcinoma identified by co-expression analysis. Oncol Rep.
39:525–536. 2018.PubMed/NCBI
|
|
19
|
Budczies J, Klauschen F, Sinn BV, Győrffy
B, Schmitt WD, Darb-Esfahani S and Denkert C: Cutoff finder: A
comprehensive and straightforward web application enabling rapid
biomarker cutoff optimization. PLoS One. 7:e518622012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lotia S, Montojo J, Dong Y, Bader GD and
Pico AR: Cytoscape app store. Bioinformatics. 29:1350–1351. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu X, Deng Y, Huang L, Feng B and Liao B:
A co-expression modules based gene selection for cancer
recognition. J Theor Biol. 362:75–82. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Deng SP, Zhu L and Huang DS: Mining the
bladder cancer-associated genes by an integrated strategy for the
construction and analysis of differential co-expression networks.
BMC Genomics. 16 (Suppl 3):S42015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Montero JC, Esparís-Ogando A, Re-Louhau
MF, Seoane S, Abad M, Calero R, Ocaña A and Pandiella A: Active
kinase profiling, genetic and pharmacological data define mTOR as
an important common target in triple-negative breast cancer.
Oncogene. 33:148–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pelicano H, Zhang W, Liu J, Hammoudi N,
Dai J, Xu RH, Pusztai L and Huang P: Mitochondrial dysfunction in
some triple-negative breast cancer cell lines: Role of mTOR pathway
and therapeutic potential. Breast Cancer Res. 16:4342014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hatem R, El Botty R, Chateau-Joubert S,
Servely JL, Labiod D, de Plater L, Assayag F, Coussy F, Callens C,
Vacher S, et al: Targeting mTOR pathway inhibits tumor growth in
different molecular subtypes of triple-negative breast cancers.
Oncotarget. 7:48206–48219. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Massihnia D, Galvano A, Fanale D, Perez A,
Castiglia M, Incorvaia L, Listì A, Rizzo S, Cicero G, Bazan V, et
al: Triple negative breast cancer: Shedding light onto the role of
pi3k/akt/mtor pathway. Oncotarget. 7:60712–60722. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Crown J, O'Shaughnessy J and Gullo G:
Emerging targeted therapies in triple-negative breast cancer. Ann
Oncol. 23 (Suppl 6):vi56–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Paolicchi E, Crea F, Farrar WL, Green JE
and Danesi R: Histone lysine demethylases in breast cancer. Crit
Rev Oncol Hematol. 86:97–103. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
McGrath J and Trojer P: Targeting histone
lysine methylation in cancer. Pharmacol Ther. 150:1–22. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li LX, Zhou JX, Calvet JP, Godwin AK,
Jensen RA and Li X: Lysine methyltransferase SMYD2 promotes triple
negative breast cancer progression. Cell Death Dis. 9:3262018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Copeland RA: Molecular pathways: Protein
methyltransferases in cancer. Clin Cancer Res. 19:6344–6350. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
McCabe MT, Mohammad HP, Barbash O and
Kruger RG: Targeting histone methylation in cancer. Cancer J.
23:292–301. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Habib JG and O'Shaughnessy JA: The
hedgehog pathway in triple-negative breast cancer. Cancer Med.
5:2989–3006. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
O'Toole SA, Beith JM, Millar EK, West R,
McLean A, Cazet A, Swarbrick A and Oakes SR: Therapeutic targets in
triple negative breast cancer. J Clin Pathol. 66:530–542. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jamdade VS, Sethi N, Mundhe NA, Kumar P,
Lahkar M and Sinha N: Therapeutic targets of triple negative breast
cancer: A review. Br J Pharmacol. 172:4228–4237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Di Mauro C, Rosa R, D'Amato V, Ciciola P,
Servetto A, Marciano R, Orsini RC, Formisano L, De Falco S,
Cicatiello V, et al: Hedgehog signalling pathway orchestrates
angiogenesis in triple-negative breast cancers. Br J Cancer.
116:1425–1435. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Seitz S, Buchholz S, Schally AV, Weber F,
Klinkhammer-Schalke M, Inwald EC, Perez R, Rick FG, Szalontay L,
Hohla F, et al: Triple negative breast cancers express receptors
for LHRH and are potential therapeutic targets for cytotoxic
LHRH-analogs, AEZS 108 and AEZS 125. BMC Cancer. 14:8472014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Buchholz S, Seitz S, Schally AV, Engel JB,
Rick FG, Szalontay L, Hohla F, Krishan A, Papadia A, Gaiser T, et
al: Triple-negative breast cancers express receptors for
luteinizing hormone-releasing hormone (LHRH) and respond to LHRH
antagonist cetrorelix with growth inhibition. Int J Oncol.
35:789–796. 2009.PubMed/NCBI
|
|
40
|
Föst C, Duwe F, Hellriegel M, Schweyer S,
Emons G and Gründker C: Targeted chemotherapy for triple-negative
breast cancers via LHRH receptor. Oncol Rep. 25:1481–1487.
2011.PubMed/NCBI
|
|
41
|
Buchholz S, Seitz S, Engel JB, Montero A,
Ortmann O, Perez R, Block NL and Schally AV: Search for novel
therapies for triple negative breast cancers (TNBC): Analogs of
luteinizing hormone-releasing hormone (LHRH) and growth
hormone-releasing hormone (GHRH). Horm Mol Biol Clin Investig.
9:87–94. 2012.PubMed/NCBI
|
|
42
|
Kwok CW, Treeck O, Buchholz S, Seitz S,
Ortmann O and Engel JB: Receptors for luteinizing hormone-releasing
hormone (GnRH) as therapeutic targets in triple negative breast
cancers (TNBC). Target Oncol. 10:365–373. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhai X, Xue Q, Liu Q, Guo Y and Chen Z:
Colon cancer recurrenceassociated genes revealed by WGCNA
coexpression network analysis. Mol Med Rep. 16:6499–6505. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shay JW, Reddel RR and Wright WE:
Cancer. Cancer and telomeres-an ALTernative to telomerase.
Science. 336:1388–1390. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Subhawong AP, Heaphy CM, Argani P, Konishi
Y, Kouprina N, Nassar H, Vang R and Meeker AK: The alternative
lengthening of telomeres phenotype in breast carcinoma is
associated with HER-2 overexpression. Mod Pathol. 22:1423–1431.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Napier CE, Huschtscha LI, Harvey A, Bower
K, Noble JR, Hendrickson EA and Reddel RR: ATRX represses
alternative lengthening of telomeres. Oncotarget. 6:16543–16558.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Watson LA, Goldberg H and Bérubé NG:
Emerging roles of ATRX in cancer. Epigenomics. 7:1365–1378. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Marinoni I, Kurrer AS, Vassella E, Dettmer
M, Rudolph T, Banz V, Hunger F, Pasquinelli S, Speel EJ and Perren
A: Loss of DAXX and ATRX are associated with chromosome instability
and reduced survival of patients with pancreatic neuroendocrine
tumors. Gastroenterology. 146:453–460.e5. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Qadeer ZA, Harcharik S, Valle-Garcia D,
Chen C, Birge MB, Vardabasso C, Duarte LF and Bernstein E:
Decreased expression of the chromatin remodeler ATRX associates
with melanoma progression. J Invest Dermatol. 134:1768–1772. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yang CY, Liau JY, Huang WJ, Chang YT,
Chang MC, Lee JC, Tsai JH, Su YN, Hung CC and Jeng YM: Targeted
next-generation sequencing of cancer genes identified frequent TP53
and ATRX mutations in leiomyosarcoma. Am J Transl Res. 7:2072–2081.
2015.PubMed/NCBI
|