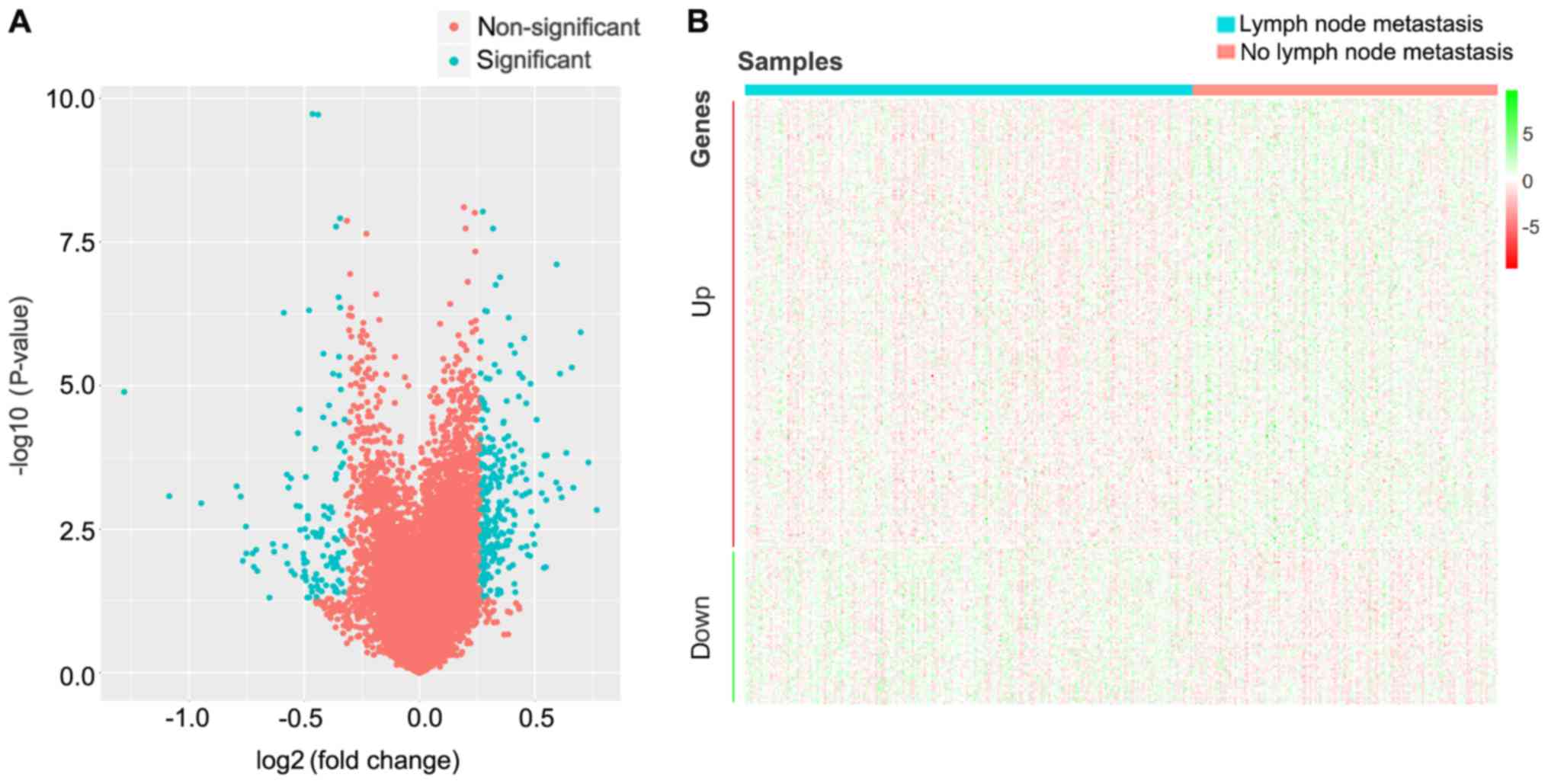
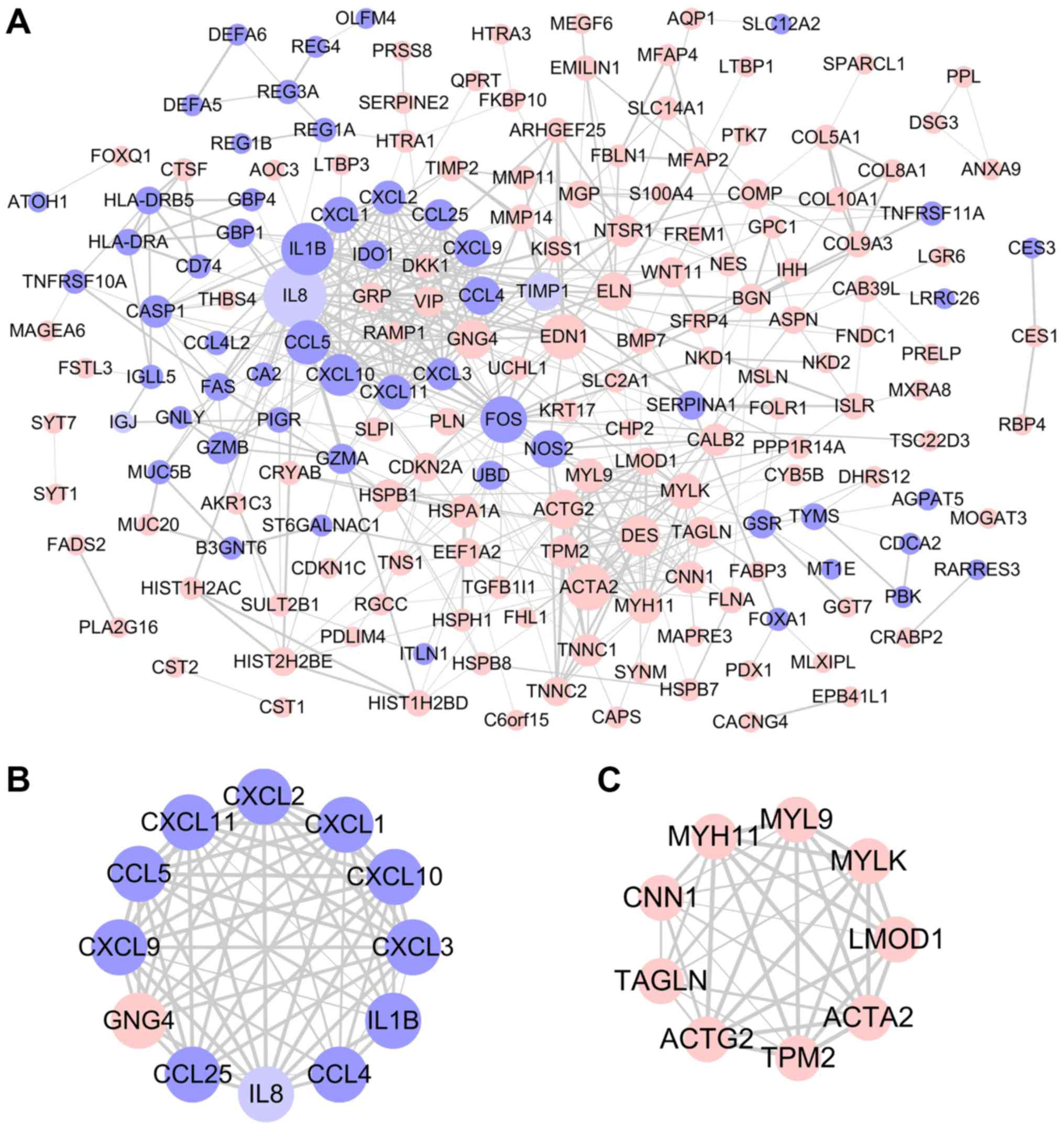
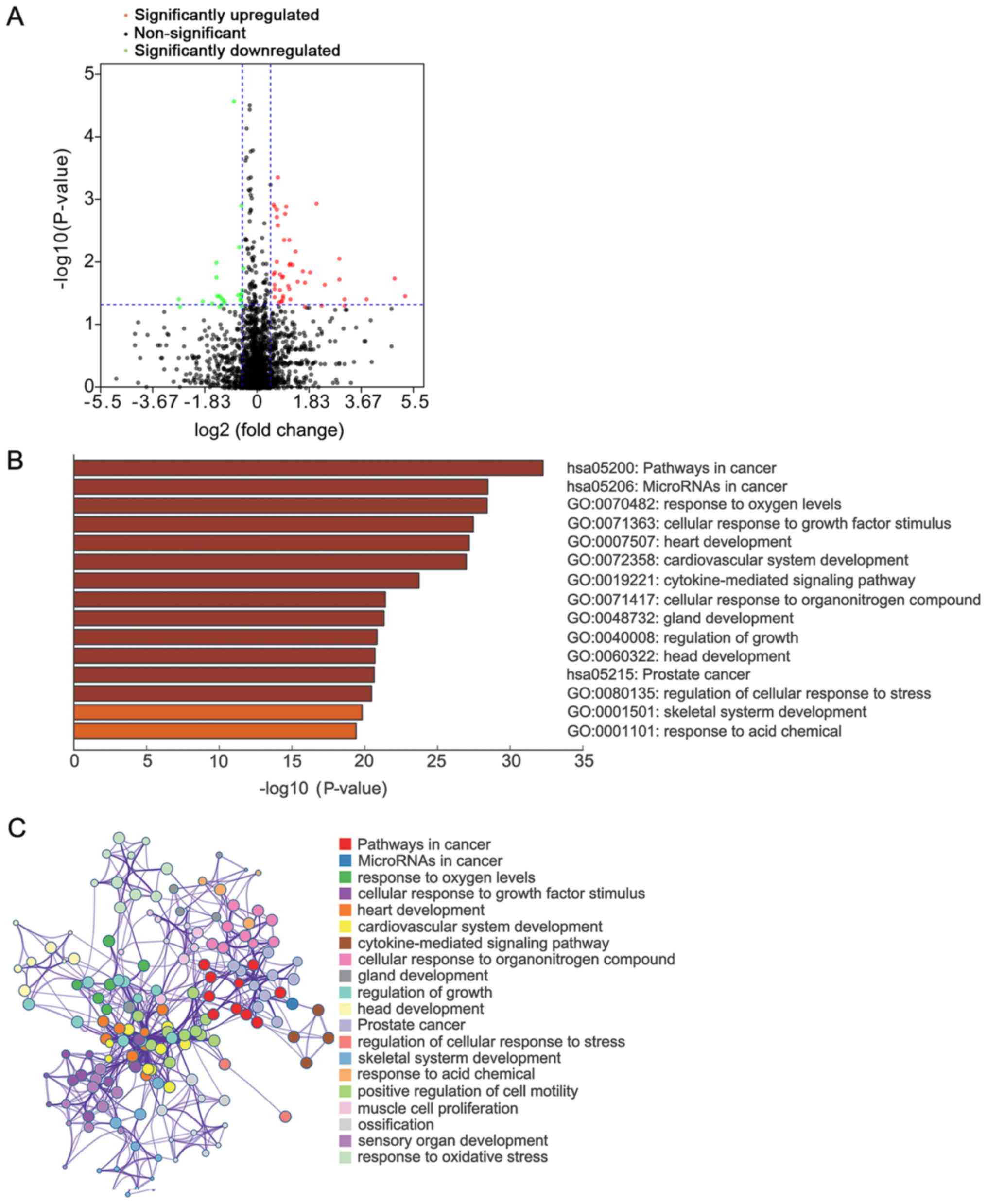
|
1
|
Liu K, Yao H, Wen Y, Zhao H, Zhou N, Lei S
and Xiong L: Functional role of a long non-coding RNA
LIFR-AS1/miR-29a/TNFAIP3 axis in colorectal cancer resistance to
pohotodynamic therapy. Biochim Biophys Acta Mol Basis Dis.
1864:2871–2880. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stintzing S, Tejpar S, Gibbs P, Thiebach L
and Lenz HJ: Understanding the role of primary tumour localisation
in colorectal cancer treatment and outcomes. Eur J Cancer.
84:69–80. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shinagawa T, Tanaka T, Nozawa H, Emoto S,
Murono K, Kaneko M, Sasaki K, Otani K, Nishikawa T, Hata K, et al:
Comparison of the guidelines for colorectal cancer in Japan, the
USA and Europe. Ann Gastroenterol Surg. 2:6–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang K, Zhang X, Cai Z, Zhou J, Cao R,
Zhao Y, Chen Z, Wang D, Ruan W, Zhao Q, et al: A novel class of
microRNA-recognition elements that function only within open
reading frames. Nat Struct Mol Biol. 25:1019–1027. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Long JM, Maloney B, Rogers JT and Lahiri
DK: Novel upregulation of amyloid-β precursor protein (APP) by
microRNA-346 via targeting of APP mRNA 5′-untranslated region:
Implications in Alzheimer's disease. Mol Psychiatry. 24:345–363.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ludwig N, Leidinger P, Becker K, Backes C,
Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stahler C, Meese E
and Keller A: Distribution of miRNA expression across human
tissues. Nucleic Acids Res. 44:3865–3877. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chipman LB and Pasquinelli AE: miRNA
targeting: Growing beyond the seed. Trends Genet. 35:215–222. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Uhlmann S, Mannsperger H, Zhang JD, Horvat
EA, Schmidt C, Kublbeck M, Henjes F, Ward A, Tschulena U, Zweig K,
et al: Global microRNA level regulation of EGFR-driven cell-cycle
protein network in breast cancer. Mol Syst Biol. 8:5702012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sells E, Pandey R, Chen H, Skovan BA, Cui
H and Ignatenko NA: Specific microRNA-mRNA regulatory network of
colon cancer invasion mediated by tissue kallikrein-related
peptidase 6. Neoplasia. 19:396–411. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hanauer DA, Rhodes DR, Sinha-Kumar C and
Chinnaiyan AM: Bioinformatics approaches in the study of cancer.
Curr Mol Med. 7:133–141. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Karagkouni D, Paraskevopoulou MD,
Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou
D, Kavakiotis I, Maniou S, Skoufos G, et al: DIANA-TarBase v8: A
decade-long collection of experimentally supported miRNA-gene
interactions. Nucleic Acids Res. 46:D239–D245. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chou CH, Shrestha S, Yang CD, Chang NW,
Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al: miRTarBase
update 2018: A resource for experimentally validated
microRNA-target interactions. Nucleic Acids Res. 46:D296–D302.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ozawa T, Kandimalla R, Gao F, Nozawa H,
Hata K, Nagata H, Okada S, Izumi D, Baba H, Fleshman J, et al: A
MicroRNA signature associated with metastasis of T1 colorectal
cancers to lymph nodes. Gastroenterology. 154:844–848. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gebert LFR and MacRae IJ: Regulation of
microRNA function in animals. Nat Rev Mol Cell Biol. 20:21–37.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sakai E, Fukuyo M, Ohata K, Matsusaka K,
Doi N, Mano Y, Takane K, Abe H, Yagi K, Matsuhashi N, et al:
Genetic and epigenetic aberrations occurring in colorectal tumors
associated with serrated pathway. Int J Cancer. 138:1634–1644.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ma Y, Peng J, Liu W, Zhang P, Huang L, Gao
B, Shen T, Zhou Y, Chen H, Chu Z, et al: Proteomics identification
of desmin as a potential oncofetal diagnostic and prognostic
biomarker in colorectal cancer. Mol Cell Proteomics. 8:1878–1890.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Arentz G, Chataway T, Price TJ, Izwan Z,
Hardi G, Cummins AG and Hardingham JE: Desmin expression in
colorectal cancer stroma correlates with advanced stage disease and
marks angiogenic microvessels. Clin Proteomics. 8:162011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Traicoff JL, De Marchis L, Ginsburg BL,
Zamora RE, Khattar NH, Blanch VJ, Plummer S, Bargo SA, Templeton
DJ, Casey G and Kaetzel CS: Characterization of the human polymeric
immunoglobulin receptor (PIGR) 3′UTR and differential expression of
PIGR mRNA during colon tumorigenesis. J Biomed Sci. 10:792–804.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Agesen TH, Sveen A, Merok MA, Lind GE,
Nesbakken A, Skotheim RI and Lothe RA: ColoGuideEx: A robust gene
classifier specific for stage II colorectal cancer prognosis. Gut.
61:1560–1567. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Y, Kuscu C, Banach A, Zhang Q,
Pulkoski-Gross A, Kim D, Liu J, Roth E, Li E, Shroyer KR, et al:
miR-181a-5p inhibits cancer cell migration and angiogenesis via
downregulation of matrix metalloproteinase-14. Cancer Res.
75:2674–2685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Przybyla L, Muncie JM and Weaver VM:
Mechanical control of epithelial-to-mesenchymal transitions in
development and cancer. Annu Rev Cell Dev Biol. 32:527–554. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gonzalez-Avila G, Sommer B, Mendoza-Posada
DA, Ramos C, Garcia-Hernandez AA and Falfan-Valencia R: Matrix
metalloproteinases participation in the metastatic process and
their diagnostic and therapeutic applications in cancer. Crit Rev
Oncol Hematol. 137:57–83. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Weng MT, Tsao PN, Lin HL, Tung CC, Change
MC, Chang YT, Wong JM and Wei SC: Hes1 increases the invasion
ability of colorectal cancer cells via the STAT3-MMP14 pathway.
PLoS One. 10:e01443222015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Andarawewa KL, Motrescu ER, Chenard MP,
Gansmuller A, Stoll I, Tomasetto C and Rio MC: Stromelysin-3 is a
potent negative regulator of adipogenesis participating to cancer
cell-adipocyte interaction/crosstalk at the tumor invasive front.
Cancer Res. 65:10862–10871. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Doll D, Keller L, Maak M, Boulesteix AL,
Siewert JR, Holzmann B and Janssen KP: Differential expression of
the chemokines GRO-2, GRO-3, and interleukin-8 in colon cancer and
their impact on metastatic disease and survival. Int J Colorectal
Dis. 25:573–581. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yamanaka R, Abe E, Sato T, Hayano A and
Takashima Y: Secondary intracranial tumors following radiotherapy
for pituitary adenomas: A systematic review. Cancers (Basel);
9(pii): pp. E1032017, PubMed/NCBI
|
|
31
|
Jeon M, You D, Bae SY, Kim SW, Nam SJ, Kim
HH, Kim S and Lee JE: Dimerization of EGFR and HER2 induces breast
cancer cell motility through STAT1-dependent ACTA2 induction.
Oncotarget. 8:50570–50581. 2016.PubMed/NCBI
|
|
32
|
Wu Y, Liu ZG, Shi MQ, Yu HZ, Jiang XY,
Yang AH, Fu XS, Xu Y, Yang S, Ni H, et al: Identification of ACTG2
functions as a promoter gene in hepatocellular carcinoma cells
migration and tumor metastasis. Biochem Biophys Res Commun.
491:537–544. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pace-Asciak CR: Hepoxilins in cancer and
inflammation-Use of hepoxilin antagonists. Cancer Metastasis Rev.
30:493–506. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gagnaire A, Nadel B, Raoult D, Neefjes J
and Gorvel JP: Collateral damage: Insights into bacterial
mechanisms that predispose host cells to cancer. Nat Rev Microbiol.
15:109–128. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
McDermott AJ, Falkowski NR, McDonald RA,
Frank CR, Pandit CR, Young VB and Huffnagle GB: Role of
interferon-γ and inflammatory monocytes in driving colonic
inflammation during acute clostridium difficile infection in mice.
Immunology. 150:468–477. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Long AG, Lundsmith ET and Hamilton KE:
Inflammation and colorectal cancer. Curr Colorectal Cancer Rep.
13:341–351. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Raskov H, Burcharth J and Pommergaard HC:
Linking gut microbiota to colorectal cancer. J Cancer. 8:3378–3395.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bischoff SC, Barbara G, Buurman W,
Ockhuizen T, Schulzke JD, Serino M, Tilg H, Watson A and Wells JM:
Intestinal permeability-A new target for disease prevention and
therapy. BMC Gastroenterol. 14:1892014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bultman SJ: Emerging roles of the
microbiome in cancer. Carcinogenesis. 35:249–255. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nieto MA, Huang RY, Jackson RA and Thiery
JP: Emt: 2016. Cell. 166:21–45. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fang Y, Zhang L, Li Z, Li Y, Huang C and
Lu X: MicroRNAs in DNA damage response, carcinogenesis, and
chemoresistance. Int Rev Cell Mol Biol. 333:1–49. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kenneth NS and Rocha S: Regulation of gene
expression by hypoxia. Biochem J. 414:19–29. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ortmann B, Druker J and Rocha S: Cell
cycle progression in response to oxygen levels. Cell Mol Life Sci.
71:3569–3582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang H, Jiang H, Van De Gucht M and De
Ridder M: Hypoxic radioresistance: Can ROS be the key to overcome
it. Cancers (Basel). 11(pii): E1122019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Inokuma T, Haraguchi M, Fujita F,
Torashima Y, Eguchi S and Kanematsu T: Suppression of reactive
oxygen species develops lymph node metastasis in colorectal cancer.
Hepatogastroenterology. 59:2480–2483. 2012.PubMed/NCBI
|
|
47
|
Lin YC, Chen CC, Chen WM, Lu KY, Shen TL,
Jou YC, Shen CH, Ohbayashi N, Kanaho Y, Huang YL and Lee H: LPA1/3
signaling mediates tumor lymphangiogenesis through promoting CRT
expression in prostate cancer. Biochim Biophys Acta Mol Cell Biol
Lipids. 1863:1305–1315. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Alvarez SW, Sviderskiy VO, Terzi EM,
Papagiannakopoulos T, Moreira AL, Adams S, Sabatini DM, Birsoy K
and Possemato R: NFS1 undergoes positive selection in lung tumours
and protects cells from ferroptosis. Nature. 551:639–643. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lizarbe MA, Calle-Espinosa J,
Fernandez-Lizarbe E, Fernandez-Lizarbe S, Robles MA, Olmo N and
Turnay J: Colorectal cancer: From the genetic model to
posttranscriptional regulation by noncoding RNAs. Biomed Res Int.
2017:73542602017. View Article : Google Scholar : PubMed/NCBI
|