Introduction
Esophageal cancer, including squamous cell carcinoma
and adenocarcinoma, affected half a million people worldwide in
2014, and causes a fatal outcome in the majority of cases (1,2). The
incidence rate of esophageal cancer has increased dramatically over
the past 40 years (2). Alcohol
consumption and tobacco use are major risk factors (3–5).
Untreated gastroesophageal reflux disease usually progresses to
Barrett's esophagus, which is 50–100 times more likely to develop
into cancer (1). Obesity can also
increase the risk of developing esophageal cancer, via low-grade
inflammation (2). Due to the lack of
serosa in the esophageal wall and extensive lymphatic drainage to
intrathorax and intra-abdominal lymph nodes, esophageal cancer can
spread rapidly into the neck and thorax (2). Advanced stages of esophageal cancer can
also spread hematogenously (2).
Long non-coding (lnc)RNA colon cancer-associated
transcript 2 (CCAT2) was first identified as a region within the
8q24 chromosomal location containing a single nucleotide
polymorphism and expressed in microsatellite-stable colorectal
cancer. This was subsequently identified as a biomarker for poor
prognosis and metastasis in esophageal squamous cell carcinoma
(ESCC), gastric cancer, bladder cancer, non-small cell lung cancer
(NSCLC), small cell lung cancer (SCLC), glioma, ovarian cancer,
breast cancer and others (6–9). A number of studies have explored the
effect of lncRNA CCAT2 on cancer, using small interfering (si)RNA
to inhibit its expression in various cancer cell lines (such as
HuCCT1, CCLP1, DU-145, 22RV1, MDA-MB-231 and MCF-7) (6–13). The
results demonstrated that the silencing of CCAT2 inhibits
proliferation and invasion of NSCLC, SCLC, glioma, ovarian cancer,
bladder cancer, breast cancer and even tamoxifen-resistant breast
cancer cells (14–17). These studies indirectly or directly
demonstrated that overexpression of lncRNA CCAT2 promotes cell
proliferation, migration and invasion, and inhibits cell apoptosis,
implying that it exerts a tumor-promoting function in many cancer
types (10–17). The more extensive studies found that
CCAT2 promotes proliferation and metastasis via the Wnt signaling
pathway and epithelial-mesenchymal transition (10–13).
Moreover, it has been reported to regulate cancer cell metabolism
via alternative splicing of glutaminase by selecting the poly(A)
site in intron 14 of the precursor mRNA (18).
LncRNA CCAT2 was found to be associated with ESCC by
in silico analysis and subsequent clinical studies (4,19). These
studies showed that CCAT2 was highly expressed in cancer tissue and
was associated with smoking (4,19). Lymph
node metastasis, advanced lymph node metastasis and Myc
amplification were also associated with high lncRNA CCAT2
expression (19). Furthermore, the
expression of this lncRNA was positively correlated with Myc
amplification and progression of cancer (19).
To investigate the molecular mechanism of lncRNA
CCAT2 function in esophageal cancer, the expression of lncRNA CCAT2
in esophageal cancer cells and its association with proliferation
and metastasis were investigated in the present study. The results
from the current study may provide a theoretical basis for new
treatment options for esophageal cancer.
Materials and methods
Cell culture
Normal human esophageal epithelial cells (HEEC; cat.
no. BNCC337729; BeNa Culture Collection; www.bnbio.com) and human esophageal cancer cell lines
KYSE150 (cat. no. BNCC342590; BeNa Culture Collection; www.bnbio.com), Eca-109 (cat. no. BNCC337687), EC9706
(cat. no. BNCC339892) and TE-1 (cat. no. BNCC100151) (all BeNa
Culture Collection) were cultured in RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc.) containing 10% FBS (Sigma-Aldrich;
Merck KGaA) under conditions of 37°C and 5% CO2 in a
cell incubator (Thermo Fisher Scientific, Inc.). Growth phase cells
(80% confluence) were used for subsequent experiments.
Grouping
Based on the expression level of lncRNA CCAT2 in the
four esophageal cancer cell lines, the Eca-109 cell line was
selected for further experiments because it exhibited a higher
expression level of lncRNA CCAT2 than the other three cell lines.
The cells were divided into 5 groups: i) Control, no treatment; ii)
negative control group (si-NC), scramble sequence transfected; iii)
lncRNA CCAT2-silenced group (si-CCAT2), transfection with siRNA
sequences targeting CCAT2; iv) Wnt pathway inhibitor (FH535) group,
10 µM FH535 treatment (cat. no. HY-15721; MedChemExpress); and v)
CCAT2 silencing and inhibitor group (si-CCAT2 + FH535),
transfection with siRNA sequence targeting CCAT2 and 10 µM FH535
treatment. The esophageal cancer Eca-109 cells were digested and
passaged with 0.25% trypsin (Invitrogen; Thermo Fisher Scientific,
Inc.). Cells (2×105) were inoculated on a 6-well plate.
After 24 h, when the cells reached 30–50% confluence, 10 µl/250 µl
siRNA was transfected with Lipofectamine® 2000
Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
A further ~24 h later, the medium containing the mixture was
preplaced with fresh medium containing 10% FBS. Cell transfection
was assessed by reverse transcription-quantitative PCR (RT-qPCR).
The sequences of the control and lncRNA CCAT2 siRNAs were as
follows: si-CCAT2 sense strand, 5′-GCCUGUAGGAAGAGUCAAATT-3′;
si-CCAT2 antisense strand, 5′-UUUGACUCUUCCUACAGGCTT-3′; si-NC sense
strand, 5′-UUCUCCGAACGUGUCACGUTT-3′; and si-NC antisense strand,
5′-ACGUGACACGUUCGGAGAATT-3′.
RT-qPCR
Total RNA was extracted from each cell line using
the Total RNA Extraction kit (Invitrogen; Thermo Fisher Scientific,
Inc.), and RNA was reverse-transcribed into cDNA using SuperScript
III Reverse Transcriptase (Thermo Fisher Scientific, Inc.). SYBR
Green PCR kit (Qiagen, Inc.) and Mastercycler® Ep
Realplex2 (Eppendorf) were used for RT-qPCR. The RT-qPCR was
performed using 2 µl cDNA as a template under the following
conditions: 95°C for 10 min, 95°C for 15 sec and 60°C for 1 min for
40 cycles. GAPDH was used as the internal reference and the
relative expression of lncRNA CCAT2 was calculated by the
2−ΔΔCq method (20). The
following primer pairs were used: CCAT2 forward,
5′-AGACAGTGCCAGCCAACC-3′ and reverse, 5′-TGCCAAACCCTTCCCTTA-3′; and
GAPDH forward, 5′-AATGGACAACTGGTCGTGGAC-3′ and reverse,
5′-CCCTCCAGGGGATCTGTTTG-3′.
MTT assay
Cells (~100 µl; 1×104 cells/ml) were
inoculated on a 96-well flat-bottomed culture plate and cultured
for 24, 48, 72 and 96 h. MTT (100 µg) was added and incubated at
37°C for 4 h. The supernatant was dissolved with 200 µl DMSO
(Sigma-Aldrich; Merck KGaA). Optical density (OD) of each well was
measured at a wavelength of 490 nm using a plate reader (Bio-Rad
Laboratories). The relative absorbance of each group was calculated
by the following equation:
Relative
absorbance=ODExperiment-ODBlankODControl-ODBlank
Wound-healing assay
Cells (~1.0 ml; 3×105 cells/ml) were
plated in each well (6-well plate). When cells reached 100%
confluence within 24 h, a 10-µl pipette tip was used to scratch the
monolayer of cells. The cells were washed with PBS twice to remove
suspended cells and serum-free medium was gently added. The
cell-free wound area was photographed with an inverted microscope
(Olympus Corporation) and the results were analyzed by ImageJ 1.46r
software (National Institutes of Health). The percentage of the
wound size at 24 h relative to the 0 h time point of each treatment
group was calculated.
Transwell assay
Matrigel mixture (~50 µl) was added to the upper
chamber of Transwell inserts (EMD Millipore) and incubated at 37°C
for 30 min. Cells (2×105 cells/ml) were resuspended in
serum-free DMEM (Gibco; Thermo Fisher Scientific, Inc.). A total of
~100 µl cell suspension was added into the upper compartment of the
chamber, and 600 µl of 10% FBS-DMEM culture media was added to the
lower chamber. Membranes were collected after 24 h incubation,
fixed in 5% glutaraldehyde at 4°C for 30 min, stained with 0.5%
crystal violet at room temperature for 20 min and observed under a
light microscope (Olympus Corporation) at ×200 magnification.
Images of nine random fields were captured for analysis of the
number of invading cells.
Flow cytometry
Cells were digested by trypsin and collected by
centrifugation at 11,180 × g for 5 min at 4°C. The cells were
washed twice with pre-cooled sterile PBS solution at 4°C. Cells
[~195 µl; 1×106 cells in 250 µl 1X binding buffer
(Beyotime Institute of Biotechnology)] were incubated with 5 µl of
FITC-labeled annexin-V (Beyotime Institute of Biotechnology) for 3
min. Then, 10 µl propidium iodide solution (20 µg/ml) was added and
incubated in the dark for 10 min at room temperature. Binding
buffer (~400 µl 1X solution) was added prior to analysis using a
flow cytometer (Gallios; Beckman Coulter, Inc.) and Cell Quest 5.1
software (BD Biosciences). The cell numbers at different stages
were recorded and the ratio of apoptotic cells was defined as the
ratio of cells in quadrants 2 and 4 to total cells.
Immunocytochemistry
Cells, cultured for 24 h, were washed with PBS two
to three times and fixed with paraformaldehyde for 30 min. PBS was
used to wash the cells two to three times prior to adding 0.5%
Triton X-100 in PBS and incubated for 20 min. This was followed by
two to three PBS washes before 3% H2O2 was
added for 15–20 min. Cells were then washed with PBS two to three
times and 5% bovine serum albumin (Beyotime Institute of
Biotechnology) was added and incubated at 37°C for 30 min. The
primary antibodies, including rabbit anti-human anti-β-catenin
(1:200; cat. no. ab6302) or anti-PCNA (1:100; cat. no. ab15497)
(both Abcam), were added and incubated at 37°C for 30 min. After
two to three PBS washes, horseradish peroxidase-conjugated goat
anti-rabbit IgG (1:1,000; cat. no. ABIN101988; antibodies-online
GmBH) was added at 37°C for 30 min. Cells were washed with PBS two
to three times before streptavidin-biotin complex (Beyotime
Institute of Biotechnology) was added and incubated at 37°C for 30
min. Following three washes with PBS, 3,3′-diaminobenzidine was
added, followed by incubation with hematoxylin at room temperature
for 2 min and washing with ethanol at increasing concentrations
(70% alcohol for 1 min, 80% alcohol for 1 min, 95% alcohol for 2
min and absolute ethanol for 4 min). Observations were performed at
×400 magnification under a light microscope (Olympus Corporation)
and three to five fields of each section were randomly selected for
analysis.
Western blotting
Cells were lysed in RIPA buffer (Beyotime Institute
of Biotechnology) and the protein concentration was measured by the
bicinchoninic acid (Pierce™ BCA Protein Assay kit; Thermo Fisher
Scientific, Inc.). Each protein sample (~40 µg) was separated on a
SDS-PAGE 10% gel (Mini-Protean-3 type; Bio-Rad Laboratories) and
then transferred to a PVDF membrane (Merck KGaA) for 30 min. The
membrane was blocked with 5% non-fat dry milk in TBST solution for
1 h. TBST with 3% bovine serum albumin was used to dilute each of
the antibodies, including rabbit anti-human anti-Bax (cat. no.
ab53154), anti-APC (ab15270, Abcam), anti-cyclin D1 (cat. no.
ab226977), anti-c-Myc (cat. no. ab39688) and β-actin (cat. no.
ab8227) (all 1:1,000) polyclonal antibodies, and goat anti-rabbit
IgG (1:2,000; cat. no. ab6721) (all Abcam). The membrane was
incubated with primary antibody overnight at 4°C. Following
incubation with horseradish peroxidase-labeled goat anti-rabbit
secondary antibody (1:1,000; cat. no. ABIN101988; Antibodies
Online) at room temperature for 1 h, the ECL system (Thermo Fisher
Scientific, Inc.) was used to detect the signals. The expression
levels were quantified by ImageJ 1.46r software, and β-actin was
used as an internal control.
Statistical analysis
Each experiment was repeated at least three times.
SPSS 19.0 (IBM Corp.) was used to analyze all the data, which are
expressed as the mean ± SD. One-way analysis of variance was used
for analysis of data among groups. Least-Significant Difference was
used for post hoc analysis. P<0.05 was considered to indicate s
statistically significant difference.
Results
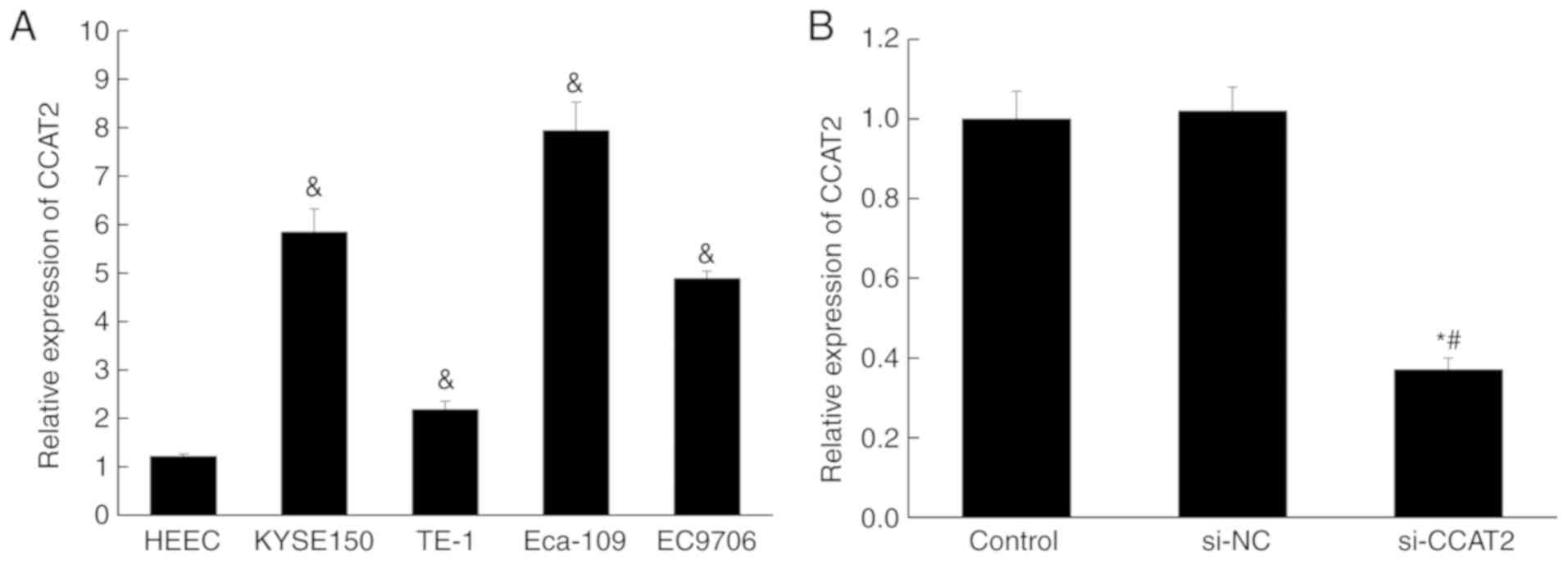
Expression of lncRNA CCAT2 is higher
in esophageal cancer cell lines
The expression of lncRNA CCAT2 in four esophageal
cancer cell lines (Eca-109, EC9706, KYSE150 and TE-1) and one
normal esophageal epithelial cell (HEEC) was examined by RT-qPCR.
The expression was significantly higher in all esophageal cancer
cell lines compared with that in HEEC (P<0.05; Fig. 1A). Subsequently, siRNA was used to
silence lncRNA CCAT2 in Eca-109 cells. The RT-qPCR data showed no
off-target effect occurring, and the knockdown of lncRNA CCAT2 was
successfully performed (Fig.
1B).
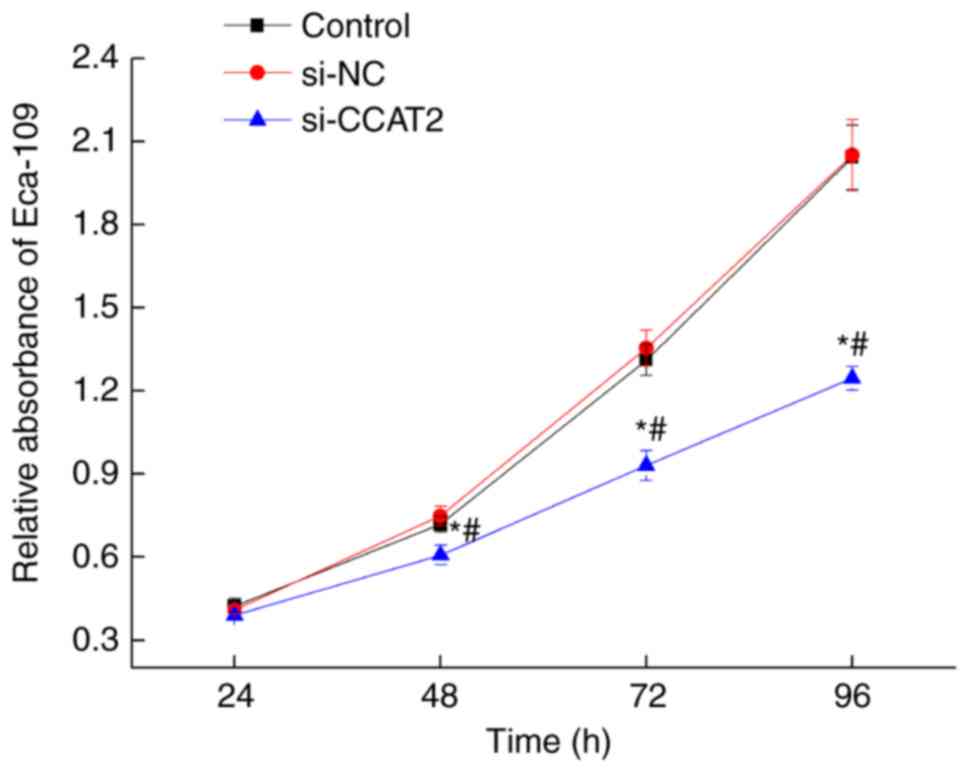
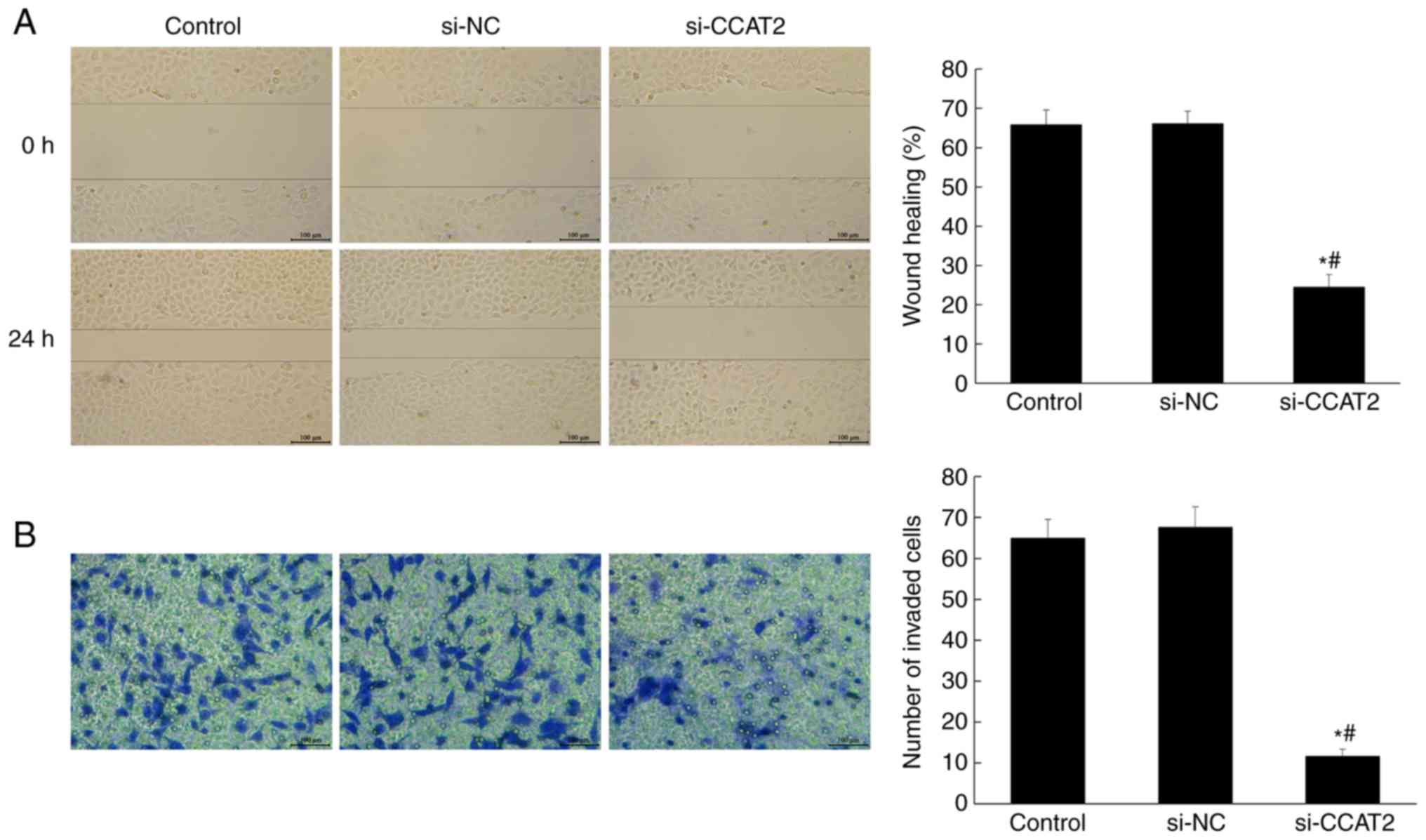
Inhibition of lncRNA CCAT2 attenuates
proliferation, migration and invasion of esophageal cancer
cells
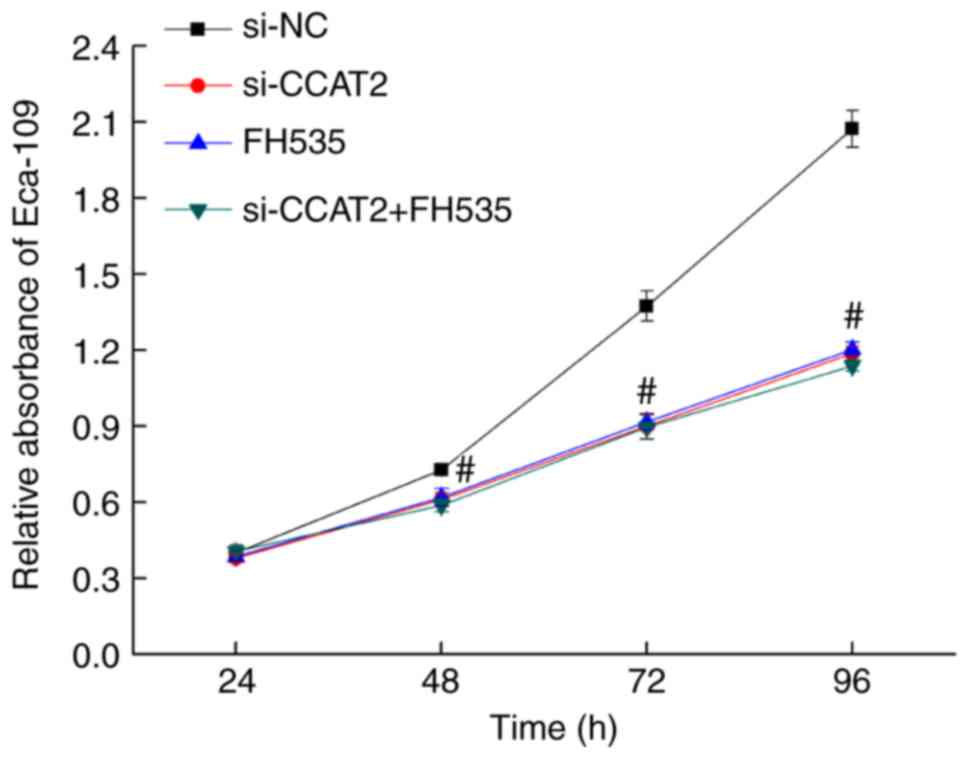
After 24, 48, 72, and 96 h of culture following
transfection with si-CCAT2, proliferation decreased significantly
in the si-CCAT2 group compared with the control and si-NC groups
(P<0.05; Fig. 2). To further
investigate the effect of lncRNA CCAT2 on the metastasis of
esophageal cancer cells, the wound-healing and Transwell assays
were performed. As shown in Fig. 3,
the cell migration and invasion in the si-CCAT2 group were
significantly suppressed compared with the control and si-NC groups
(P<0.05).
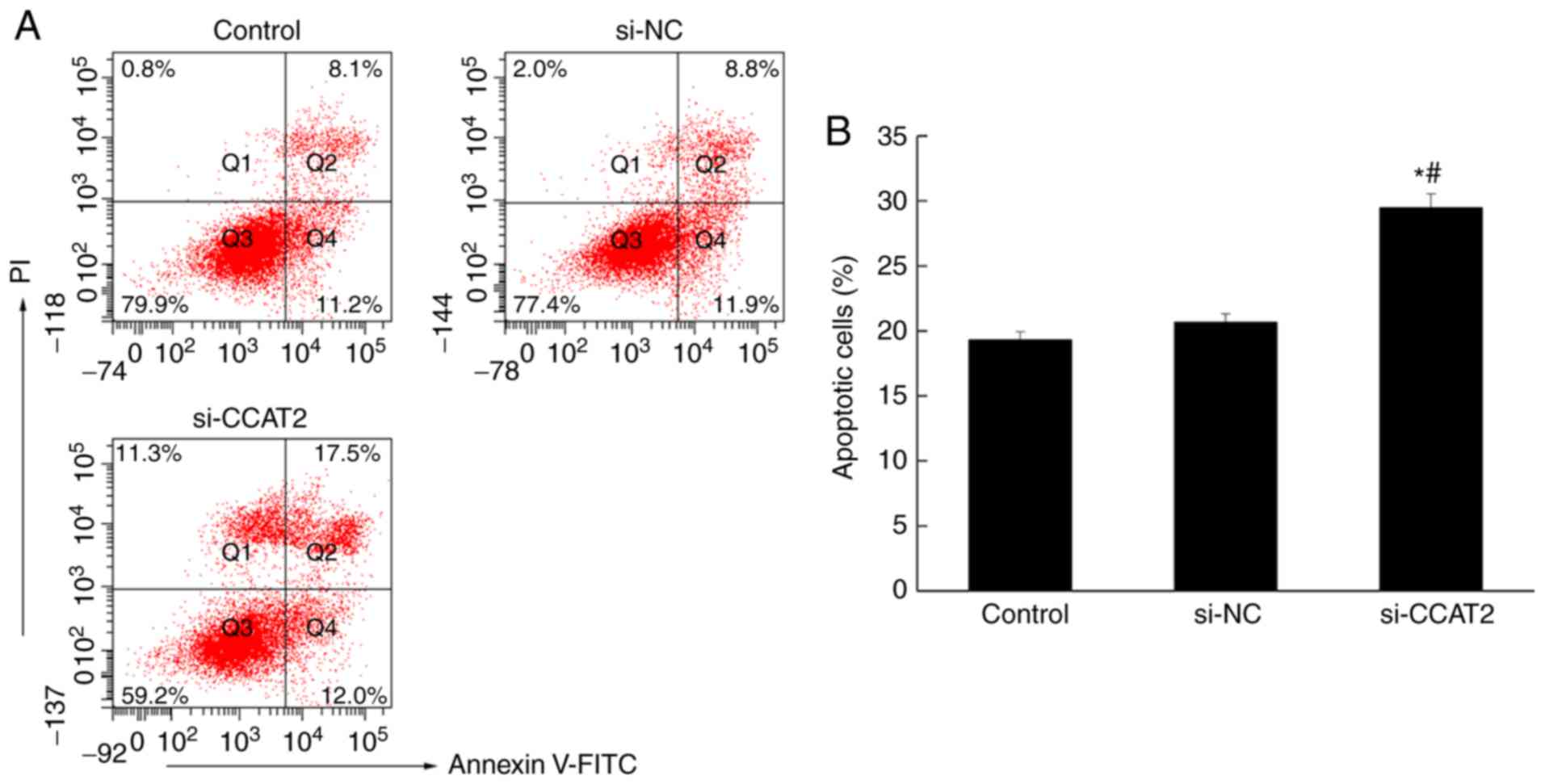
Inhibition of lncRNA CCAT2 promotes
apoptosis in esophageal cancer cells
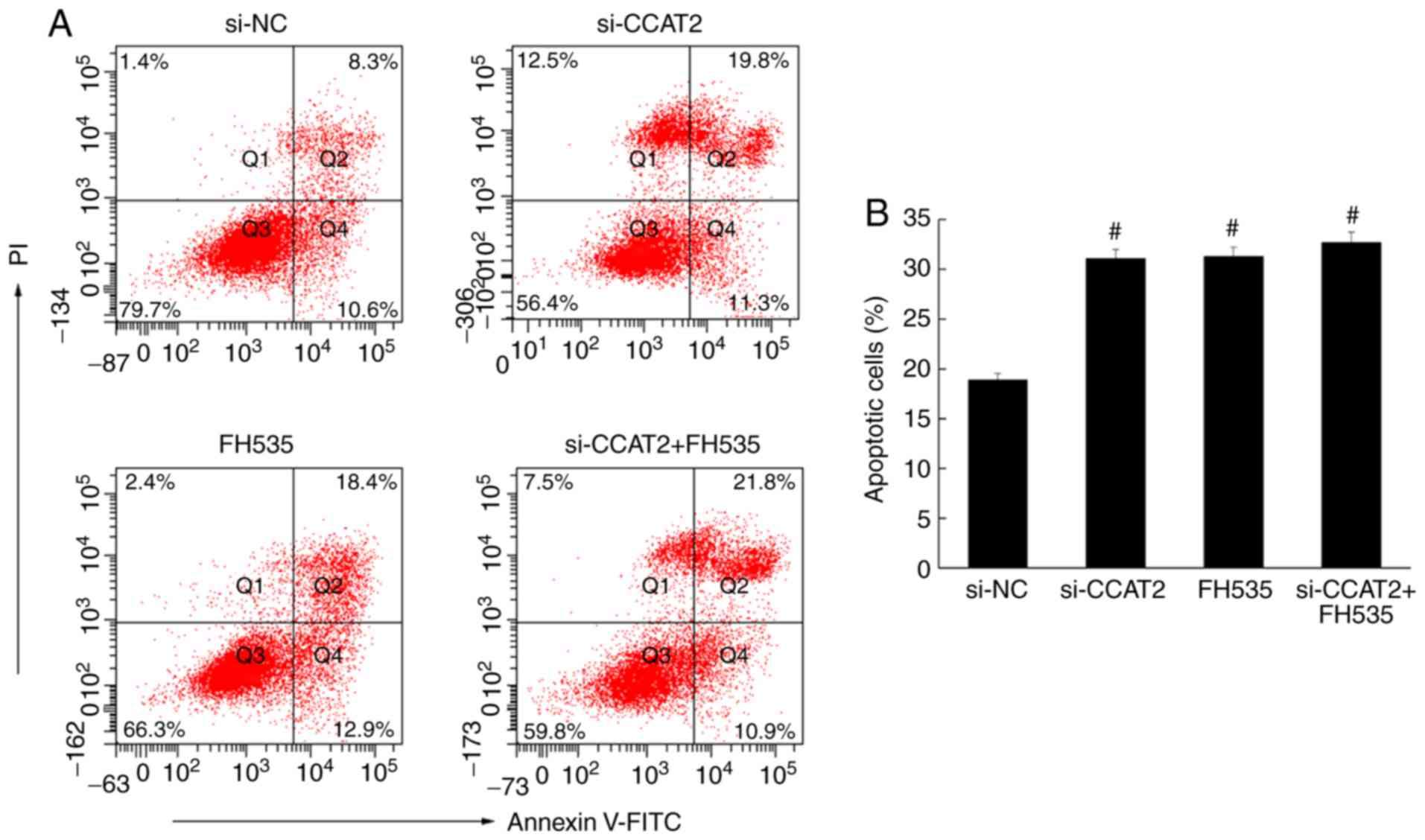
Flow cytometry was used to analyze apoptosis in
esophageal cancer cells, following annexin V staining. The number
of apoptotic cells in the si-CCAT2 group was significantly higher
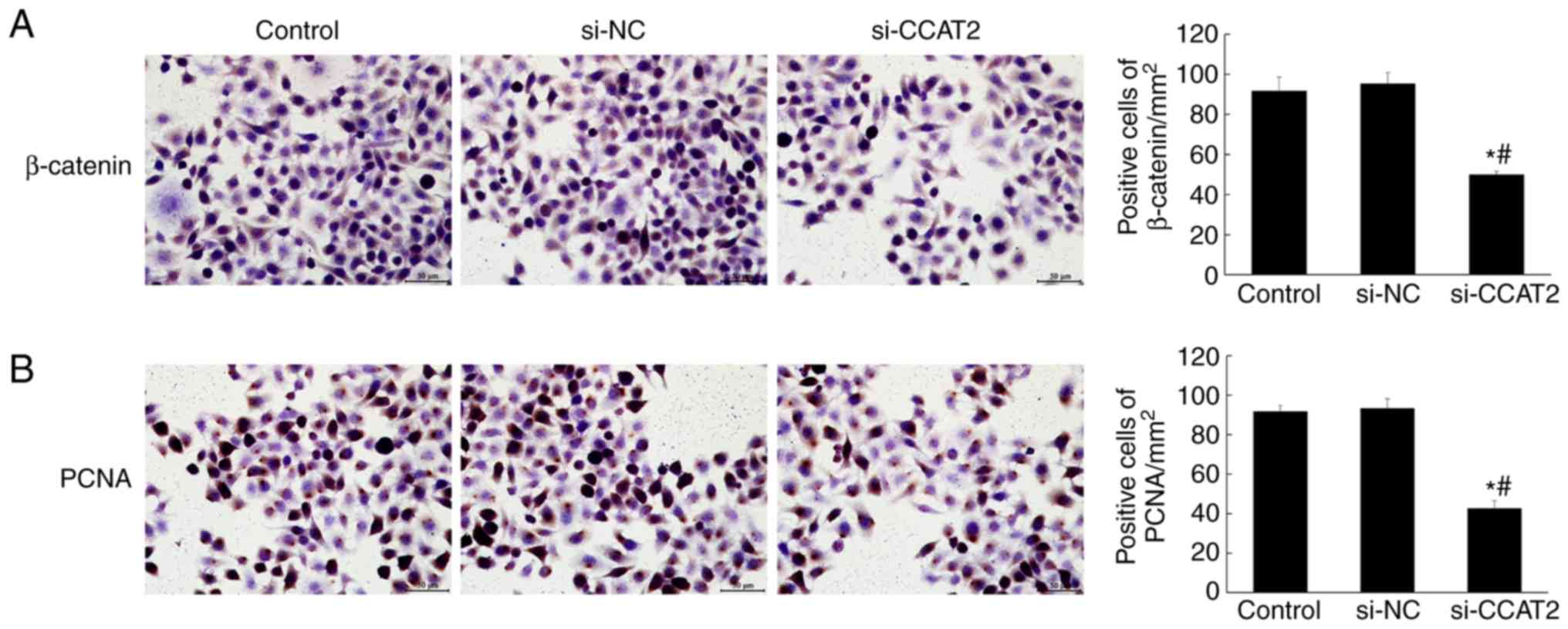
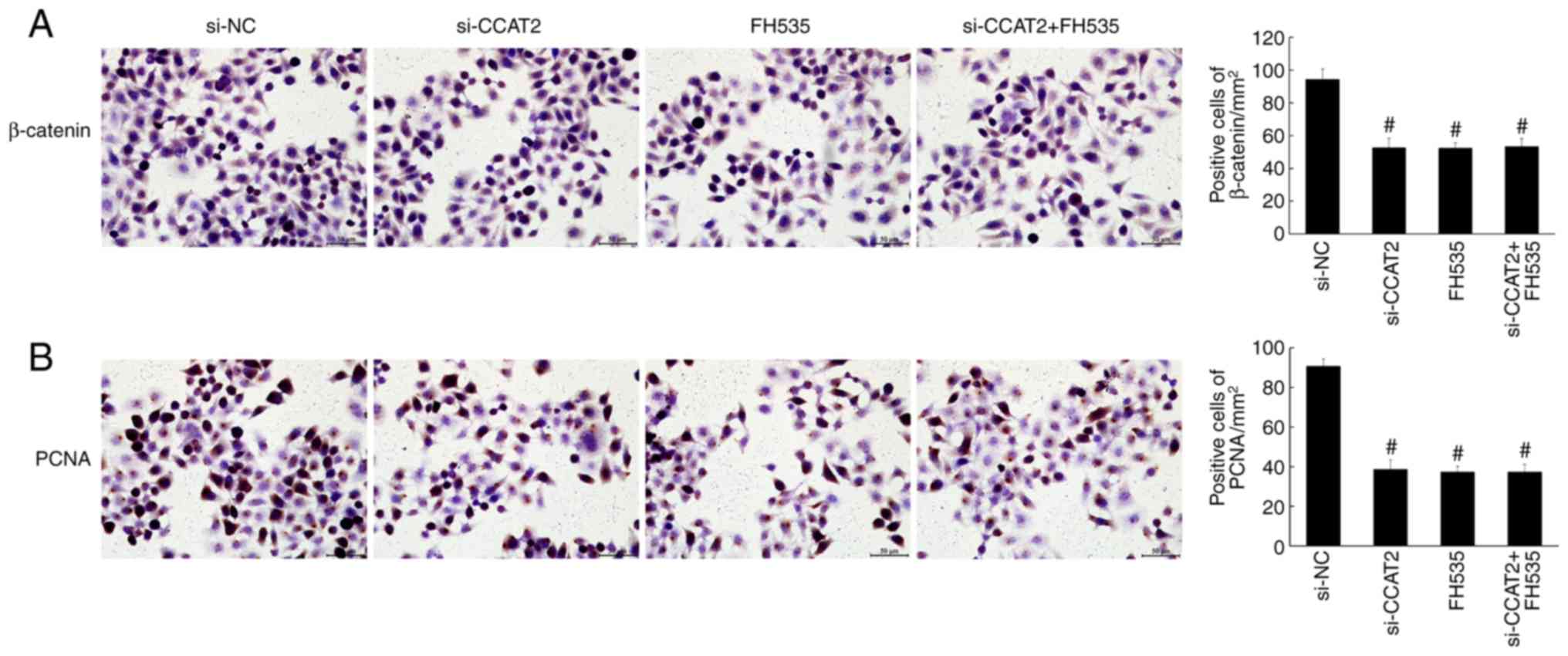
compared with the control and si-NC groups (P<0.05; Fig. 4). Similarly, the immunohistochemistry
staining showed significantly decreased expression of β-catenin and
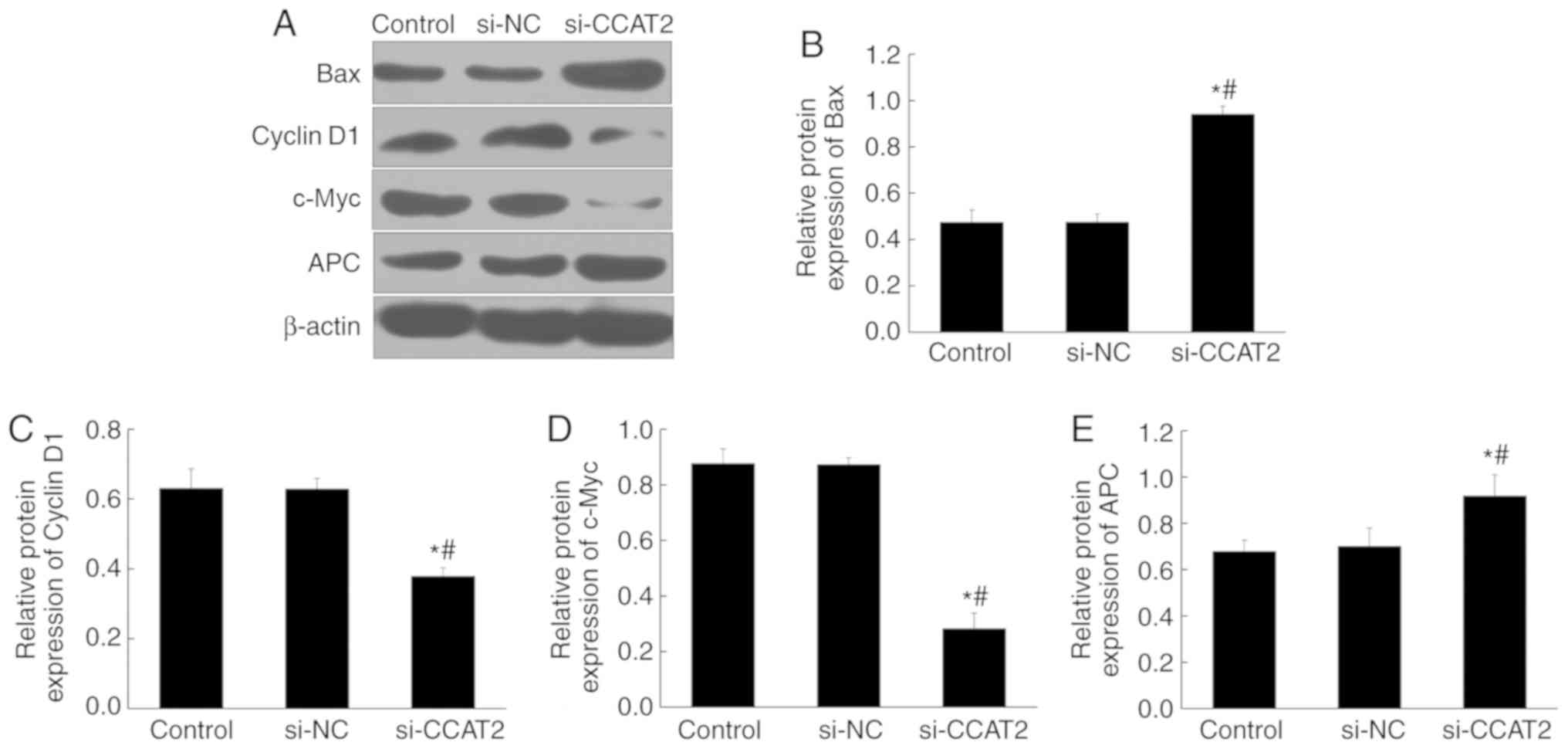
PCNA in the si-CCAT2 group (P<0.05; Fig. 5). Protein expression, determined by
western blotting, showed significant upregulation of Bax and APC in
the si-CCAT2 group compared with the control and si-NC groups,
whereas cyclin D1 and c-Myc proteins were significantly
downregulated (P<0.05; Fig.
6).
LncRNA CCAT2 and Wnt signaling
pathway
To further analyze how lncRNA CCAT2 functions in
esophageal cancer cells, the effect of the Wnt signaling pathway on
the cell proliferation, invasion and metastasis was investigated,
using a Wnt signaling inhibitor (FH535). No differences in the
effects on apoptosis, proliferation, migration and invasion of
Eca-109 cells were observed between the si-CCAT2 + FH535 group and
the si-CCAT2 or FH535 groups (Figs.
7–9). Expression of β-catenin,
PCNA, Bax, APC, cyclin D1 and c-Myc were also similar among these
groups (Figs. 10 and 11).
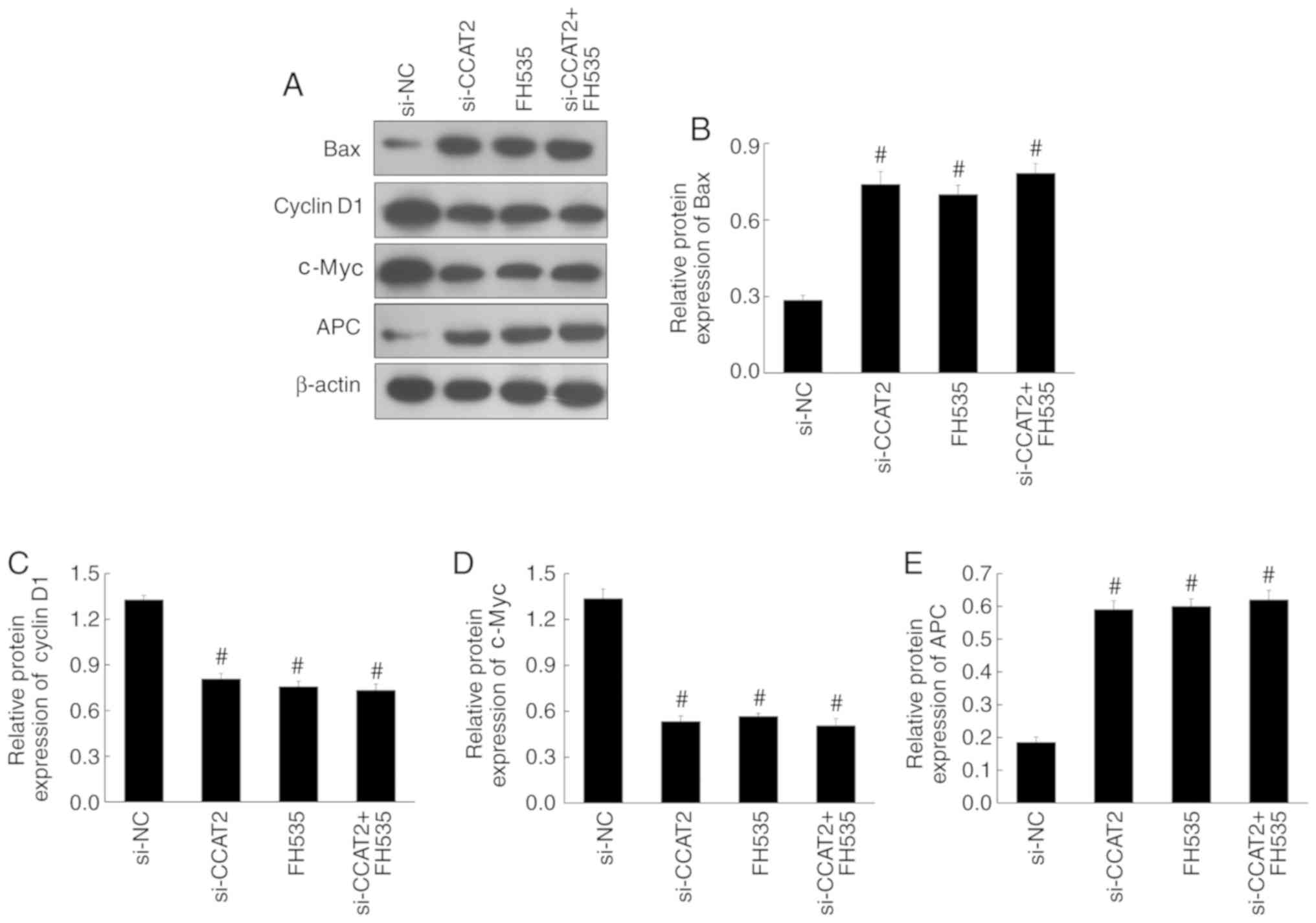 | Figure 11.Effect of the Wnt pathway inhibitor
(FH535) on the expression levels of pro-apoptotic and Wnt target
proteins in Eca-109 cells. (A) The expression of Bax, cyclin D1,
c-Myc and APC by western blotting. The relative protein expression
of (B) Bax, (C) cyclin D1, (D) c-Myc and (E) APC in the si-CCAT2,
FH535 and si-CCAT2 + FH535 groups, compared with the si-NC group.
#P<0.05 vs. si-NC. APC, adenomatous polyposis coli;
CCAT2, colon cancer-associated transcript 2; si-CCAT2, small
interfering RNA targeting CCAT2; si-NC, small interfering RNA
negative control. |
Discussion
β-catenin can bind T-cell factor (TCF) family
DNA-binding proteins and translocate to the nucleus. This complex
regulates cellular differentiation and proliferation (21–24). Wnt
signaling molecules can activate the β-catenin/TCF complex and
induce gene expression (21–23). This signaling pathway has been
identified to serve a major role in in certain cancer types
including highly malignant carcinoma such as esophageal cancer
(21,25–27). It
is believed that this pathway plays a critical role in the
maintenance and self-renewal of cancer stem cells, which leads to
malignant tumors and poor prognosis (22,25). Wnt
signaling is positively correlated with higher ratio of cancer stem
cells and advanced clinical stages of esophageal carcinoma
(25). Other genes, such as Dapper
homolog 2, naked cuticle homolog 2, microRNA (miR)-942,
transcription factor Sox17 and miR-141, can modulate the Wnt
signaling pathway and affect tumorigenesis in esophageal cells
(22,23,26,28).
Human papillomavirus can lead to esophageal cancer via the miR-125b
and Wnt/β-catenin signaling pathway (27).
Therefore, the inhibition of the Wnt/β-catenin
pathway can serve as a potential therapeutic approach for cancer
(29–31). FH535 is an inhibitor of β-catenin,
which inhibits cyclin D1 and survivin and decreases the
proliferation of human colorectal cancer cells, liver cancer stem
cells, hepatocellular carcinoma cells and HepG2 cells (24,32–34).
FH535 enhances imatinib-induced apoptosis, decreases S phase cells,
arrests the cell cycle and suppresses the proliferation of cancer
cells by targeting the Wnt signaling pathway (30,32–34).
The present study demonstrated that esophageal
cancer cell lines had higher expression levels of lncRNA CCAT2
compared with normal esophageal cells. To further explore the
function of lncRNA CCAT2, si-CCAT2 was constructed and tested in
esophageal cancer Eca-109 cells. The results showed that si-CCAT2
decreased the cell proliferation. Furthermore, the population of
apoptotic cells, quantified by annexin V staining, increased
significantly following knockdown of lncRNA CCAT2. This
downregulation also significantly decreased cell migration and
invasion. To elucidate the molecular mechanism of lncRNA CCAT2, a
Wnt inhibitor (FH535) was used to study the associated signaling
pathway. FH535 and lncRNA CCAT2 equally elicited suppressive
effects on proliferation, migration and invasion, suggesting that
lncRNA may function via the Wnt pathway.
LncRNA CCAT2 has been demonstrated to be involved in
the Wnt pathway in a number of cancer types, and one of its
functions is to modulate the Wnt pathway (35–38). For
example, downregulating CCAT2 suppresses the transcriptional
activity of Wnt/β-catenin signaling pathway and the combination of
siCCAT2 and FH535 synergistically inhibits Wnt signaling in breast
cancer (39). The present study
demonstrates an association between lncRNA CCAT2 and Wnt signaling
pathway in esophageal cancer.
In the present study, only one esophageal cancer
cell line (Eca-109) was used to study the effects and mechanism of
lncRNA CCAT2. Therefore, a limitation in the present findings is
that they may not apply to other esophageal cancer cell lines or
different esophageal cancer types. In addition, the present results
showed that the expression level of lncRNA CCAT2 was different in
the four esophageal cancer cell lines, which was not investigated
any further in this study. Furthermore, more detailed studies of
the regulatory effect of CCAT2 on the Wnt pathway should be
conducted.
In the future, the correlation between the
expression level of lncRNA CCAT2 and the proliferation and
metastasis of the four esophageal cancer cell lines will be
explored. In addition, the hypothesis tested in the present study
will be applied in an in vivo model. These studies will
provide more critical insights into the mechanism, for the
development of novel treatments for esophageal cancer in the
future.
In summary, the present study demonstrated that
lncRNA CCAT2 was highly expressed in esophageal cancer cells, and
its downregulation led to the inhibition of cell proliferation,
migration and invasion via the Wnt signaling pathway. The results
of the present study may provide a theoretical basis for the
development of new treatment options for esophageal cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XiW and XuW conducted all the experiments, analyzed
all the data and wrote the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bajpai M, Das KM, Lefferts J, Lisovsky M,
Mashimo H, Phillips WA, Srivastava A and To H: Molecular
epidemiology of and genetic susceptibility to esophageal cancer.
Ann N Y Acad Sci. 1325:40–48. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Napier KJ, Scheerer M and Misra S:
Esophageal cancer: A Review of epidemiology, pathogenesis, staging
workup and treatment modalities. World J Gastrointest Oncol.
6:112–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y,
Ueda J, Wei W, Inoue M and Tanaka H: Epidemiology of esophageal
cancer in Japan and China. J Epidemiol. 23:233–242. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang J, Qiu M, Xu Y, Li M, Dong G, Mao Q,
Yin R and Xu L: Long noncoding RNA CCAT2 correlates with smoking in
esophageal squamous cell carcinoma. Tumour Biol. 36:5523–5528.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Łaźniak S, Lutkowska A, Wareńczak-Florczak
Ż, Sowińska A, Tsibulski A, Roszak A, Sajdak S and Jagodziński PP:
The association of CCAT2 rs6983267 SNP with MYC expression and
progression of uterine cervical cancer in the Polish population.
Arch Gynecol Obstet. 297:1285–1292. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jing X, Liang H, Cui X, Han C, Hao C and
Huo K: Long noncoding RNA CCAT2 can predict metastasis and a poor
prognosis: A meta-analysis. Clin Chim Acta. 468:159–165. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kasagi Y, Oki E, Ando K, Ito S, Iguchi T,
Sugiyama M, Nakashima Y, Ohgaki K, Saeki H, Mimori K and Maehara Y:
The expression of CCAT2, a novel long noncoding RNA transcript, and
rs6983267 single-nucleotide polymorphism genotypes in colorectal
cancers. Oncology. 92:48–54. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lang HL, Hu GW, Zhang B, Kuang W, Chen Y,
Wu L and Xu GH: Glioma cells enhance angiogenesis and inhibit
endothelial cell apoptosis through the release of exosomes that
contain long non-coding RNA CCAT2. Oncol Rep. 38:785–798. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ozawa T, Matsuyama T, Toiyama Y, Takahashi
N, Ishikawa T, Uetake H, Yamada Y, Kusunoki M, Calin G and Goel A:
CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21
‘gene desert’, serve as important prognostic biomarkers in
colorectal cancer. Ann Oncol. 28:1882–1888. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Redis RS, Sieuwerts AM, Look MP, Tudoran
O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, et al:
CCAT2, a novel long non-coding RNA in breast cancer: Expression
study and clinical correlations. Oncotarget. 4:1748–1762. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang YJ, Liu JZ, Lv P, Dang Y, Gao JY and
Wang Y: Long non-coding RNA CCAT2 promotes gastric cancer
proliferation and invasion by regulating the E-cadherin and LATS2.
Am J Cancer Res. 6:2651–2660. 2016.PubMed/NCBI
|
|
12
|
Zheng J, Zhao S, He X, Zheng Z, Bai W,
Duan Y, Cheng S, Wang J, Liu X and Zhang G: The up-regulation of
long non-coding RNA CCAT2 indicates a poor prognosis for prostate
cancer and promotes metastasis by affecting epithelial-mesenchymal
transition. Biochem Biophys Res Commun. 480:508–514. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma Y, Hu X, Shang C, Zhong M and Guo Y:
Silencing of long non-coding RNA CCAT2 depressed malignancy of oral
squamous cell carcinoma via Wnt/β-catenin pathway. Tumour Biol.
39:10104283177176702017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L
and Yin R: CCAT2 is a lung adenocarcinoma-specific long non-coding
RNA and promotes invasion of non-small cell lung cancer. Tumour
Biol. 35:5375–5380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cai Y, He J and Zhang D: Suppression of
long non-coding RNA CCAT2 improves tamoxifen-resistant breast
cancer cells' response to tamoxifen. Mol Biol (Mosk). 50:821–827.
2016.(In Russian). PubMed/NCBI
|
|
16
|
Deng X, Zhao Y, Wu X and Song G:
Upregulation of CCAT2 promotes cell proliferation by repressing the
P15 in breast cancer. Biomed Pharmacother. 91:1160–1166. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fan YH, Fang H, Ji CX, Xie H, Xiao B and
Zhu XG: Long noncoding RNA CCAT2 can predict metastasis and poor
prognosis: A meta-analysis. Clin Chim Acta. 466:120–126. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Redis RS, Vela LE, Lu W, Ferreira de
Oliveira J, Ivan C, Rodriguez-Aguayo C, Adamoski D, Pasculli B,
Taguchi A, Chen Y, et al: Allele-specific reprogramming of cancer
metabolism by the long non-coding RNA CCAT2. Mol Cell. 61:520–534.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang X, Xu Y, He C, Guo X, Zhang J, He C,
Zhang L, Kong M, Chen B and Zhu C: Elevated expression of CCAT2 is
associated with poor prognosis in esophageal squamous cell
carcinoma. J Surg Oncol. 111:834–839. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mizushima T, Nakagawa H, Kamberov YG,
Wilder EL, Klein PS and Rustgi AK: Wnt-1 but not epidermal growth
factor induces beta-catenin/T-cell factor-dependent transcription
in esophageal cancer cells. Cancer Res. 62:277–282. 2002.PubMed/NCBI
|
|
22
|
Ge C, Wu S, Wang W, Liu Z, Zhang J, Wang
Z, Li R, Zhang Z, Li Z, Dong S, et al: miR-942 promotes cancer stem
cell-like traits in esophageal squamous cell carcinoma through
activation of Wnt/β-catenin signalling pathway. Oncotarget.
6:10964–10977. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jia Y, Yang Y, Zhan Q, Brock MV, Zheng X,
Yu Y, Herman JG and Guo M: Inhibition of SOX17 by microRNA 141 and
methylation activates the WNT signaling pathway in esophageal
cancer. J Mol Diagn. 14:577–585. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shi M, Cheng J, He Y, Jiang Z, Bodinga BM,
Liu B, Chen H and Li Q: Effect of FH535 on in vitro maturation of
porcine oocytes by inhibiting WNT signaling pathway. Anim Sci J.
89:631–639. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pang Y, Liu J, Li X, Zhang Y, Zhang B,
Zhang J, Du N, Xu C, Liang R, Ren H, et al: Nano Let7b
sensitization of eliminating esophageal cancer stemlike cells is
dependent on blockade of Wnt activation of symmetric division. Int
J Oncol. 51:1077–1088. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cao B, Yang W, Jin Y, Zhang M, He T, Zhan
Q, Herman JG, Zhong G and Guo M: Silencing NKD2 by promoter region
hypermethylation promotes esophageal cancer progression by
activating Wnt signaling. J Thorac Oncol. 11:1912–1926. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zang B, Huang G, Wang X and Zheng S:
HPV-16 E6 promotes cell growth of esophageal cancer via
downregulation of miR-125b and activation of Wnt/β-catenin
signaling pathway. Int J Clin Exp Pathol. 8:13687–13694.
2015.PubMed/NCBI
|
|
28
|
Zhang M, Linghu E, Zhan Q, He T, Cao B,
Brock MV, Herman JG, Xiang R and Guo M: Methylation of DACT2
accelerates esophageal cancer development by activating Wnt
signaling. Oncotarget. 7:17957–17969. 2016.PubMed/NCBI
|
|
29
|
Gustafson CT, Mamo T, Shogren KL, Maran A
and Yaszemski MJ: FH535 suppresses osteosarcoma growth in vitro and
inhibits Wnt signaling through tankyrases. Front Pharmacol.
8:2852017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Suknuntha K, Thita T, Togarrati PP,
Ratanachamnong P, Wongtrakoongate P, Srihirun S, Slukvin I and
Hongeng S: Wnt signaling inhibitor FH535 selectively inhibits cell
proliferation and potentiates imatinib-induced apoptosis in myeloid
leukemia cell lines. Int J Hematol. 105:196–205. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wen D, Liao T, Ma B, Qu N, Shi RL, Lu ZW,
Wang YL, Wei WJ and Ji QH: Downregulation of CSN6 attenuates
papillary thyroid carcinoma progression by reducing Wnt/β-catenin
signaling and sensitizes cancer cells to FH535 therapy. Cancer Med.
7:285–296. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu J, Li G, Liu D and Liu J: FH535
inhibits the proliferation of HepG2 cells via downregulation of the
Wnt/β-catenin signaling pathway. Mol Med Rep. 9:1289–1292. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gedaly R, Galuppo R, Daily MF, Shah M,
Maynard E, Chen C, Zhang X, Esser KA, Cohen DA, Evers BM, et al:
Targeting the Wnt/β-catenin signaling pathway in liver cancer stem
cells and hepatocellular carcinoma cell lines with FH535. PLoS One.
9:e992722014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen Y, Rao X, Huang K, Jiang X, Wang H
and Teng L: FH535 inhibits proliferation and motility of colon
cancer cells by targeting Wnt/β-catenin signaling pathway. J
Cancer. 8:3142–3153. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Guo H, Hu G, Yang Q, Zhang P, Kuang W, Zhu
X and Wu L: Knockdown of long non-coding RNA CCAT2 suppressed
proliferation and migration of glioma cells. Oncotarget.
7:81806–81814. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang JL, Liao Y, Qiu MX, Li J and An Y:
Long non-coding RNA CCAT2 promotes cell proliferation and invasion
through regulating Wnt/β-catenin signaling pathway in clear cell
renal cell carcinoma. Tumour Biol. 39:10104283177113142017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sarrafzadeh S, Geranpayeh L, Tasharrofi B,
Soudyab M, Nikpayam E, Iranpour M, Mirfakhraie R, Gharesouran J and
Ghafouri-Fard S and Ghafouri-Fard S: Expression study and clinical
correlations of MYC and CCAT2 in breast cancer patients. Iran
Biomed J. 21:303–311. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang B, Liu M, Zhuang R, Jiang J, Gao J,
Wang H, Chen H, Zhang Z, Kuang Y and Li P: Long non-coding RNA
CCAT2 promotes epithelial-mesenchymal transition involving
Wnt/β-catenin pathway in epithelial ovarian carcinoma cells. Oncol
Lett. 15:3369–3375. 2018.PubMed/NCBI
|
|
39
|
Cai Y, He J and Zhang D: Long noncoding
RNA CCAT2 promotes breast tumor growth by regulating the Wnt
signaling pathway. Onco Targets Ther. 8:2657–2664. 2015.PubMed/NCBI
|