Introduction
TP53, the first tumor suppressor gene to be
identified, acts as the guardian of the genome and is involved in
the regulation of several essential cell processes, including, but
not limited to, cell cycle regulation, apoptosis, cell
differentiation, DNA repair and blood vessel formation (1,2). It is
the most frequently mutated gene across a large spectrum of
different types of cancer, including lung adenocarcinoma and lung
squamous cell carcinoma, with a mutation rate of ~50% (2–4). Under
normal conditions, TP53 is rapidly degraded; however, upon
cellular stress, it is activated and stabilized, resulting in
protein accumulation in the nucleus (5,6). The
activation of the TP53 signaling pathway has been
demonstrated to lead to DNA damage repair and cell cycle arrest
(7,8). Mutations in TP53 have been
revealed to result in the loss of tumor-suppressor function, thus
leading to an unstable genome and downregulating apoptosis
(9). Accumulating evidence have
suggested that, in addition to eliminating the tumor suppressor
function, mutations in TP53 can also induce new functions,
including gain-of-function mutations, which can accelerate tumor
progression and metastasis (2,9,10).
TP53 mutation is observed in ~50% of patients
with non-small cell lung cancer (NSCLC), with a higher prevalence
in squamous-cell carcinoma of the lung compared with lung
adenocarcinoma (38 vs. 12%) (11,12).
These alterations can include frameshift, nonsense, silent and
missense mutations (11–13). Unlike other tumor suppressor genes,
such as APC, BRCA1 or RB transcriptional corepressor 1
(RB1) with truncating mutations being the major alteration
type, the majority of TP53 alterations are missense
mutations, accounting for more than 75% of alterations (13,14). The
majority of TP53 mutations occur in the DNA-binding region,
in exons 5–8, spanning 540 nucleotides with numerous recurring
hotspot mutations, leading to a stable protein with a significant
loss of activity (14–17). In vitro studies have shown
that wild-type (WT) p53 promotes gefitinib-induced apoptosis
(18). The prognostic and predictive
values of TP53 mutations have been investigated, however
results are conflicting; a previous study demonstrated that
non-disruptive TP53 mutations are independently correlated
with shorter OS in patients with advanced NSCLC, regardless of
epidermal growth factor receptor (EGFR) and KRAS
status (19). Another study revealed
that a shorter OS was associated with adjuvant chemotherapy in
patients presenting mutations in TP53 with NSCLC and
completely resected tumors (20). A
previous study investigating clinical outcomes of patients with
NSCLC with dual EGFR and TP53 mutations revealed
lower response rates and shorter progression-free survival (PFS) in
such patients compared with patients with EGFR mutations
(21). By contrast, other previous
studies have revealed the lack of association between TP53
mutations and OS or response to treatment (22–24). The
lack of a unifying classification system may contribute to the
controversy regarding the prognosis and predictive value of
TP53. A variety of criteria have been used to categorize
TP53 mutations, including, but not limited to, functional
effects on p53 (disruptive vs. non-disruptive) (15,17,19) and
location (‘hotspot’ exons vs. ‘non-hotspot’ exons) (14,17).
Currently, in clinical settings, all TP53
mutations have been considered equally, without major differences
among the various types of mutations. However, an increasing number
of studies suggested that the type and position of the mutation may
be important, and the present study aimed to investigate this
possibility in lung cancer. In fact, numerous studies have revealed
that the position and the type of mutation have differential
effects on prognosis (2,11,17).
Important functional differences among various mutant forms of p53
have been elucidated, including mutations in the amino-terminal
(AT) domain, the oligomerization domain (OD) and the DNA-binding
domain (DBD) (25–28). AT-domain mutations often result in
the disruption of the expression of full-length p53 (26). Alternatively, translation from the
start codon in exon 4 results in the expression of p47, which
retains the apoptotic function of p53 (26). A previous study has suggested that
sporadic human cancers with AT-domain mutations are often more
responsive to treatment (26).
Mutations occurring in the OD, which is important for the
tetramerization of p53, often behave as loss-of-function mutations
(25). Patients harboring such
mutations are less responsive to therapies that rely on
p53-mediated cytotoxic effects (25). In total, ~80% of TP53
mutations affect the DBD, encoded by exons 5–8. In addition to the
loss of functional effects, mutations in DBD can also acquire
additional oncogenic properties after the loss of the WT allele
(2,28).
In the present study, capture-based ultra-deep
targeted sequencing was performed on the plasma samples of 379
Chinese patients with advanced lung cancer to investigate clinical
outcomes associated with TP53 mutations. The TP53
mutation classification systems, based on the functional effect and
location of the mutation, were also compared.
Materials and methods
Patient selection
TP53 status was retrospectively analyzed and
its predictive and prognostic values were examined in 379 patients
with advanced lung cancer (Stage IIIB-IV) harboring at least one
classic NSCLC driver mutation. Staging of the primary lung tumor,
lymph node status and metastasis were assessed based on the
American Joint Committee on Cancer 7th edition Tumor, Node and
Metastasis (TNM) staging system of NSCLC (29). Patients (female to male ratio, 1:1.3;
median age, 56.5 years; range, 26–82 years) were treated at any of
the nine participating centers between September 2015 and October
2016. The inclusion criteria were: i) Patients diagnosed with
advanced-stage lung cancer (stage IIIB-stage IV) of any histology
harboring at least one classic NSCLC driver mutation; and ii) the
patient was treated at any of the nine participating centers
between September 2015 and October 2016. The exclusion criteria was
patients with early-stage lung cancer (stage IA-IIIA) of any
histology. Written informed consent was obtained from each patient
for participation in the present study. Capture-based targeted
sequencing was performed on the plasma samples using a panel
consisting of 168 lung cancer-associated genes, spanning 160 kb of
the human genome. The present study was approved by the Ethics
Committee of Jiangsu Province Hospital (Nanjing, China).
Next generation sequencing library
preparation and capture-based targeted DNA sequencing
Next generation sequencing was performed using a
commercial panel comprising 168 lung cancer-associated genes (Lung
Plasma; Burning Rock Biotech) in a Clinical Laboratory Improvement
Amendments-certified laboratory as previously described (30). Briefly, circulating cell-free DNA was
acquired from 4–5 ml of plasma using the QIAamp Circulating Nucleic
Acid kit (Qiagen China Co., Ltd.) according to the manufacturer's
protocol. A minimum of 50 ng of DNA is required for next-generation
sequencing library construction. A DNA library for the
next-generation sequencing experiments were constructed. Fragments
between 200 to 400 base pairs (bp) from the DNA were end-repaired,
phosphorylated and ligated with adaptors (Agencourt AMPure XP kit;
Beckman Coulter, CA, USA). Purified DNA with adaptors were then
hybridized with capture probes baits, underwent hybrid selection
with magnetic beads, and PCR amplified. The quality and the size of
the fragments were assessed using a Qubit 2.0 fluorimeter (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) with a dsDNA
high-sensitivity assay kit (Thermo Fisher Scientific, Inc.).
Indexed samples were sequenced on a Nextseq500 sequencer (Illumina,
Inc.) with pair-end reads. An average coverage of 11,816× was
reached with a limit of detection of 0.2%.
Sequence data analysis
Data were analyzed using optimized pipeline for
somatic mutation calling as previously described (30). Briefly, the sequence data were mapped
to the reference human genome (hg19) using Burrows-Wheeler Aligner
(version 0.7.10) (31). Local
alignment optimization and variant calling were performed using
Genome Analysis Tool kit (version 3.2) (32,33) and
VarScan (version 2.4.3) (34).
Variants were filtered using the VarScan fpfilter pipeline; loci
with depth <100 were filtered out. Base calling in plasma
samples required ≥8 supporting reads for single nucleotide
variations and 5 supporting reads for insertion-deletion
variations. Variants with population frequency >0.1% in the ExAC
(http://exac.broadinstitute.org/), 1,000
Genomes (35), dbSNP (https://www.ncbi.nlm.nih.gov/snp/) (36) or ESP6500SI–V2 (https://evs.gs.washington.edu/EVS/) databases were
grouped as single nucleotide polymorphisms and excluded from
further analysis. Remaining variants were annotated with ANNOVAR
(2016-02-01 release) (37) and
SnpEff (version 3.6) (38). Analysis
of DNA translocation was performed using Factera (version 1.4.3)
(39). Copy number variations (CNV)
were analyzed based on the depth of coverage data of capture
intervals using an in-house developed algorithm. The limit of
detection for CNVs was 1.5 and 2.64 for deletions and
amplifications, respectively.
Classification of TP53 mutations
Disruptive mutations, as described previously
(15), were defined as any mutation
leading to a stop codon or missense mutations occurring within the
L2-L3 loop of the DNA-binding domain, leading to a substitution
with an amino acid of a different polarity or charge group. All
other mutations were defined as non-disruptive. Hotspot exons were
defined as exons 5–8, as previously described (17).
Statistical analysis
Since the data were not equally distributed, data
are presented as the median. All statistical tests were conducted
in R version 3.3.3 (The R Foundation for Statistical Computing,
Vienna, Austria; http://www.r-project.org) and R Studio version 1.1.383
software (40), and all tests were
two-sided unless otherwise specified. Pearson's correlation test
was used to assess correlation between two continuous variables.
Fisher's exact test was used to assess the association between two
categorical variables. Survival times were illustrated by
Kaplan-Meier curves with the P-value determined by log-rank tests
or Cox regression models when a co-variant was included. All
survival analyses were adjusted for age, sex, smoking history,
stage and histology. P<0.05 was considered to indicate a
statistically significant difference.
Results
Patient characteristics
The TP53 status of 379 patients with advanced
lung cancer (Stage IIIB to IV) harboring at least one classic NSCLC
driver mutation with various histological types was assessed in the
present study. Among them, 294 patients had EGFR mutations,
24 had anaplastic lymphoma kinase (ALK) rearrangements, 14
had erb-b2 receptor tyrosine kinase 2 mutations, five had MET
Proto-Oncogene (MET) mutations, two had B-raf proto-oncogene
mutations, four had ROS proto-oncogene 1, receptor tyrosine kinase
fusions, 11 had KRAS fusions and five had ret proto-oncogene
fusions; the remaining 20 patients had dual driver mutations
(Table I). In the examined cohort,
213 (56.2%) were female and 166 (43.8%) were male patients, and 156
had a history of smoking. The median age of this cohort was 56.5
years, ranging between 26 and 82 years. The cohort primarily
consisted of adenocarcinoma (332/379), followed by squamous cell
carcinoma (SqCC) (11/379) and small cell lung cancer (10/379). A
total of 165 patients (43.5%) had not received tyrosine kinase
inhibitors (TKI) as a treatment regimen and were considered
TKI-naïve; among them, 84 were treatment-naïve and the remaining
were previously treated with chemotherapy. A total of 214 patients
(56.5%) were previously treated with TKI; among them, 184 patients
were previously treated with one line of treatment. A total of 173
patients were treated with EGFR-TKI and the remaining 11 were
treated with crizotinib, an ALK inhibitor (41). A total of 30 patients received two
lines of treatment. Among them, 24 were treated with first and
third generation EGFR-TKIs, two patients were treated with first
generation EGFR-TKIs followed by a tyrosine-protein kinase MET
inhibitor and the remaining three patients were treated with ALK
inhibitors, Crizotinib followed by ceritinib (41). Table I
summarized the detailed clinical characteristics of the cohort
investigated.
 | Table I.Patient demographics and clinical
characteristics. |
Table I.
Patient demographics and clinical
characteristics.
|
Characteristics | Value |
|---|
| Total, n | 379 |
| Sex |
|
| Male, n
(%) | 166 (43.8) |
| Female,
n (%) | 213 (56.2) |
| Age, median
(range) | 56.5 years (26–82
years) |
| Smoking
history |
|
|
Smokers, n (%) | 156 (41.2) |
|
Non-smokers, n (%) | 200 (52.8) |
| No
data, n (%) | 39 (10.3) |
| Histology |
|
|
Adenocarcinoma, n (%) | 332 (87.5) |
|
Squamous cell carcinoma, n
(%) | 11 (2.9) |
| Small
cell lung cancer, n (%) | 10
(2.64) |
| Others,
n (%) | 26 (6.9) |
| Treatment
History |
|
|
TKI-naïve, n (%) | 165 (43.5) |
| One
line of TKI treatment, n (%) | 184 (48.5) |
|
EGFR-TKIs, n (%) | 173 (94) |
|
ALK-TKIs, n (%) | 11 (6) |
| Two
lines of TKI treatment, n (%) | 30 (8) |
| 1st and
3rd EGFR-TKI, n (%) | 24
(80) |
|
EGFR-TKI and c-MET-TKI, n
(%) | 3
(10) |
|
ALK-TKIs, n (%) | 3
(10) |
| TP53 status,
n (%) |
|
| WT, n
(%) | 190 (50.1) |
|
Mutated, n (%) | 189 (49.9) |
|
Disruptive mutation, n
(%) | 84
(44.4) |
|
Non-disruptive mutation, n
(%) | 105 (55.6) |
| Exon 5,
n (%) | 48
(25.4) |
| Exon 6,
n (%) | 31
(16.4) |
| Exon 7,
n (%) | 40
(21.2) |
| Exon 8,
n (%) | 46
(24.3) |
| Driver
mutation |
|
|
EGFR, n (%) | 294 (77.6) |
|
ALK, n (%) | 24 (6.3) |
|
ERBB2, n (%) | 14 (3.7) |
|
MET, n (%) | 5
(1.3) |
|
BRAF, n (%) | 2
(0.5) |
|
ROS1, n (%) | 4
(0.1) |
|
KRAS, n (%) | 11 (2.9) |
|
RET, n (%) | 5
(1.3) |
| Dual
drivers, n (%) | 20 (5.3) |
TP53 mutation prevalence and
associations with clinical parameters
A capture-based ultra-deep targeted sequencing
analysis was performed as described in Materials and methods and
(30) on the plasma samples obtained
from 379 patients with advanced lung cancer to investigate their
TP53 status and the prognostic value of the TP53
mutations. The prevalence of TP53 mutations in the cohort
was 49.9% (189/379), which is comparable to its prevalence in the
western population (22). Among
them, 163 patients harbored mutations in hotspot exons: 48
Mutations were on exon 5, 31 on exon 6, 40 on exon 7 and 46 on exon
8 (Table I). Two patients had two
TP53 mutations (data not shown). A total of 84 patients had
disruptive mutations. The distribution of TP53 mutations is
also presented in Fig. S1. No
association between TP53 mutations and smoking history was
observed when all TP53 mutations were taken into
consideration (data not shown). The examined cohort primarily
consisted of patients with adenocarcinoma, squamous cell carcinoma
or small cell lung cancer. The percentages of TP53 mutations
were comparable in patients with adenocarcinoma (48.3%) and small
cell lung cancer (48.4%) (data not shown). All patients
investigated in the present study with small cell lung cancer
carried TP53 mutations, in line with previous studies
(42), suggesting that a significant
percentage of patients with small cell lung cancer have TP53
mutations. Next, the mutation spectra of patients with TP53
mutations were compared with patients without TP53 mutation
(Fig. S2). The present results
revealed that the most frequent mutation was EGFR in both
groups. In addition, a larger number of RB1 mutations and
MET amplifications were present in patients with TP53
mutation (Fig. S2).
The correlations between TP53 mutations,
classified according to different systems, and clinical parameters,
including but not limited to the TNM stage, smoking history and
metastatic sites were further investigated. A positive correlation
was identified between TP53 mutations and the TNM stage when
all mutations were considered collectively. Patients with
TP53 mutations were more likely to have advanced N (P=0.004,
r=0.161) and M (P=0.004, r=0.151) stages of the disease (Fig. 1A and B). A positive correlation was
also identified between liver metastasis and TP53 mutations
when all mutations were considered collectively (P=0.001, r=0.187).
In the cohort, 67.9% patients (55/81) with TP53 mutations
had liver metastasis; in contrast, 45.5% (127/279) of patients
without a TP53 mutation had liver metastasis (P=0.001)
(Fig. 1C). These trends also existed
when TP53 mutations were classified according to the
location of the mutation or functional effects on the p53 protein.
Hotspot exon and non-hotspot exon mutations demonstrated a
significant correlation with N (P=0.036 and 0.012, respectively)
and M (P=0.012 and 0.001, respectively) stage (data not shown).
When TP53 mutations were classified as disruptive or
non-disruptive mutations, TP53 disruptive mutations
exhibited a non-significant correlation with N (P=0.08) and M
(P=0.1) stages. A strong correlation was observed between liver
metastasis and TP53 mutation, regardless of the
classification system (P<0.001). Only TP53 hotspot exon
mutations were significantly correlated with bone metastasis
(P=0.032) (data not shown). Collectively, the present results
suggested that only certain types of TP53 mutations were
correlated with the clinical parameters analyzed, providing
evidence for the hypothesis that not all TP53 mutations are
equal.
Prognostic values of TP53
mutations
Conflicting findings regarding the prognostic values
of the TP53 status were reported, which can be partially
attributed to the lack of a unifying classification system
(21–24). The prognostic value of TP53
mutations was evaluated using the two aforementioned classification
systems. Patients were grouped into three groups based on their
treatment history: i) TKI-naïve (n=165); ii) previously treated
with one line of TKI (n=184); and iii) treated with ≥2 TKI (n=30).
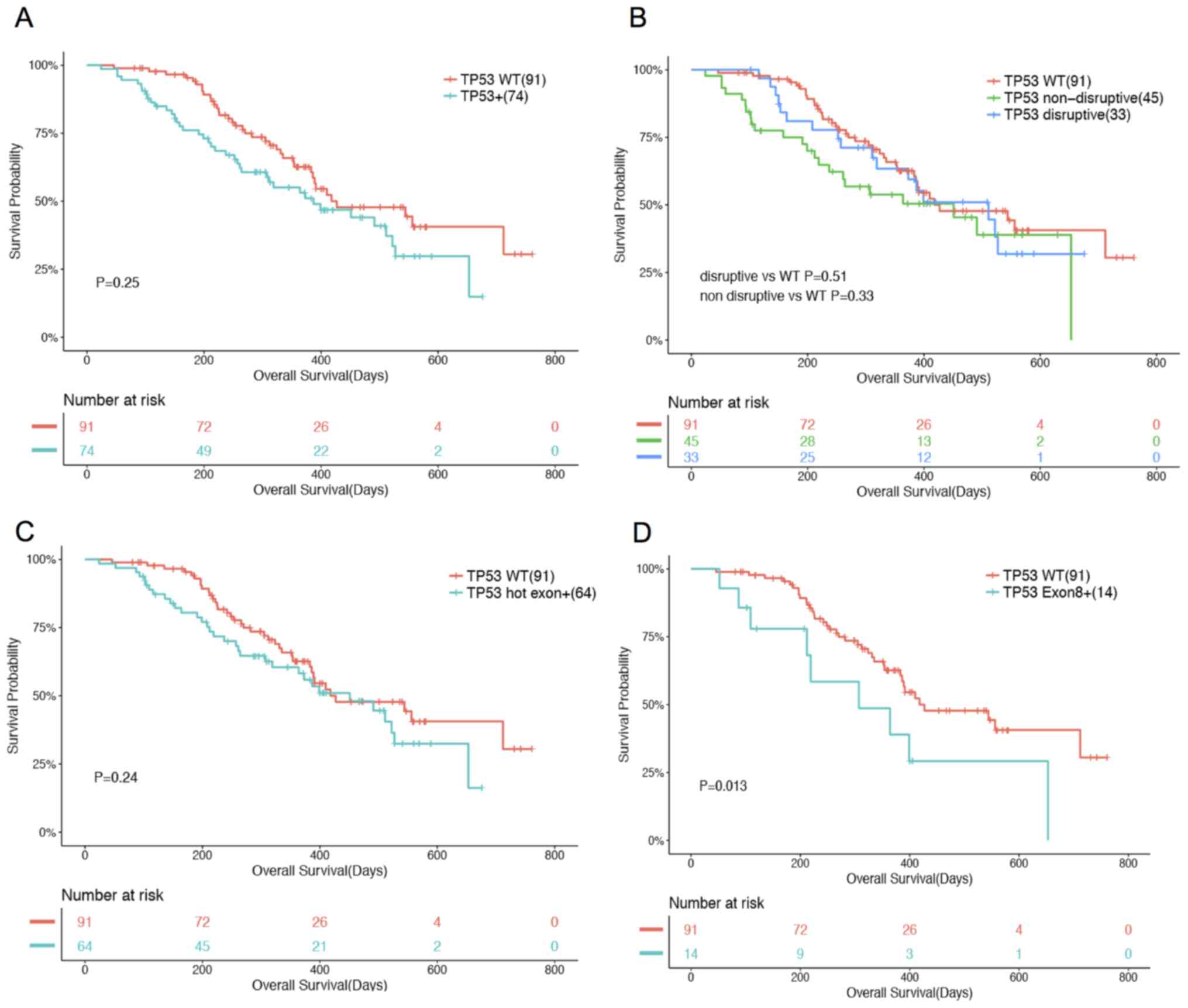
In the TKI-naïve patients, 74 patients had TP53 mutations.
Among them, 64 had mutations in hotspot exons (exons 5–8) (2,28) and
the remaining 10 had mutations in other exons. A total of 33
patients had disruptive mutations and 45 had non-disruptive
mutations. No association was observed between TP53
mutations and OS when all TP53 mutations were considered
collectively or classified according to their location (hotspot
exon vs. non-hotspot exon mutations) and functional effects on p53
protein (disruptive vs. non-disruptive; Fig. 2A-C). Notably, mutations occurring on
exon 8 were found to be associated with OS (P=0.013) when
controlling for age, sex, stage and histology. A total of 14
patients with mutations in exon 8 had a shorter median OS compared
with the remaining 91 patients who had no mutations in exon 8
(Fig. 2D).
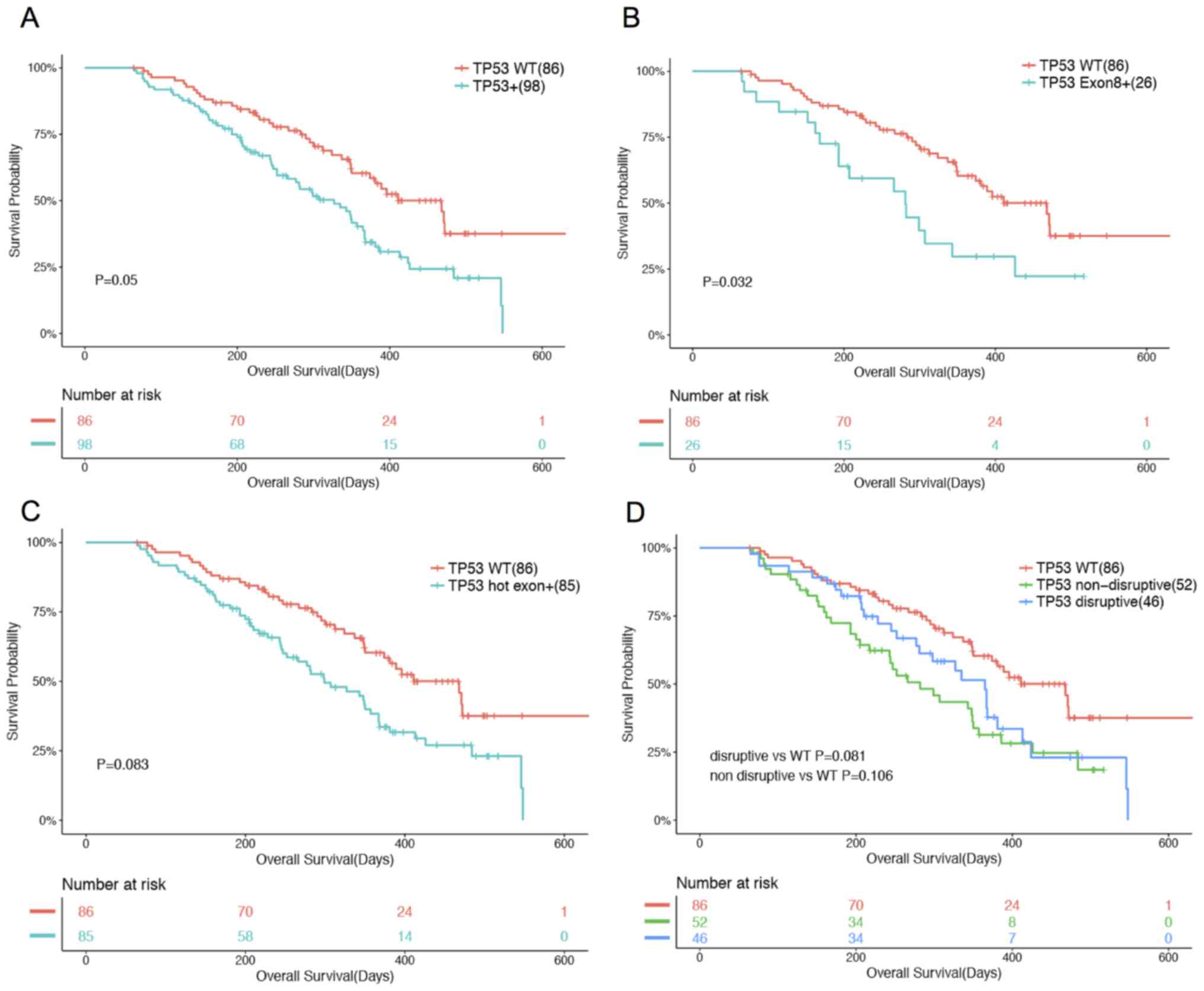
In patients previously treated with one line of TKI
treatment (n=184), an analysis revealed that TP53 status,
when all mutations were considered collectively, was found to be
marginally associated with OS (P=0.05). Patients with TP53
mutations had a shorter OS compared with patients with WT
TP53 (Fig. 3A). Such
associations were significantly enhanced when only mutations
occurring on exon 8 were considered (P=0.032; Fig. 3B). A total of 26 patients had
mutations in exon 8, including 22 missense, three frameshift and
one nonsense mutation (Fig. 3B).
However, when all hot exon mutations were considered collectively,
no association was observed (P=0.083; Fig. 3C). The same trend was observed in
TKI-naïve patients, disruptive (P=0.081) and non-disruptive
(P=0.106) mutations were not significantly associated with OS
(Fig. 3D).
In the cohort, 99 patients were undergoing
osimertinib treatment, a third-generation EGFR TKI (43). The impact of TP53 mutations on
OS in osimertinib-treated patients was subsequently analyzed. In
this cohort, 32 patients had WT TP53 and 67 patients had
TP53 mutations (data not shown). No association was observed
between TP53 status and OS, regardless of the classification
system (data not shown). In the examined cohort of patients, 62
patients possessed EGFR 19 del and 37 possessed EGFR
L858R (data not shown). No association was observed between
TP53 status and OS, regardless of classification system, in
patients harboring EGFR L858R (data not shown). In patients
harboring EGFR 19 del concurrent to T790M treated with
osimertinib, non-disruptive mutations (P=0.031) were found to be
associated with OS (Fig. 4A). A
total of 14 patients with non-disruptive TP53 mutations
exhibited a significantly shorter OS compared with patients with WT
TP53. All TP53 mutations (P=0.156), particularly
disruptive mutations (P=0.690) as well as all hotspot exon
mutations (P=0.128) including exon 8 (P=0.075) did not exhibit an
association with OS (Fig. 4).
In patients treated with two lines of TKI, the
analysis revealed that TP53 mutations, when considered
collectively, were identified to be associated with OS (P=0.037;
Fig. 5A). Mutations occurring on
exon 8 (P=0.079) as well as all hot exon mutations (P=0.052) also
exhibited an association with OS (Fig.
5B and C). In contrast, disruptive mutations (P=0.086) did not
exhibit an association with OS, whereas non-disruptive mutations
(P=0.048) exhibited a marginal association with OS (Fig. 5D). All analyses were controlled for
smoking status. Collectively, the data provided evidence supporting
the hypothesis that TP53 mutations are not equal.
Furthermore, by comparing multiple TP53 mutation
classification systems, it was identified that mutations in exon 8
may serve as prognostic biomarkers across all patients.
Discussion
In the present study, the association between
TP53 mutations, analyzed using two classification methods
(based on location and function), and OS was investigated in a
large cohort of patients with advanced lung cancer. It was
demonstrated that mutations occurring on exon 8 may serve as
prognostic biomarkers across all patients regardless of treatment
history. The present results revealed that mutations occurring in
exon 8 correlated with shorter OS in TKI-naïve and patients
previously treated with one line of TKI. Such mutations also
exhibited a slight association, although not significant, with
shorter OS in patients previously treated with two lines of
treatment. Therefore, TP53 exon 8 mutations defined a
distinct subset of patients with an unfavorable prognosis. The
association between OS and TP53 mutations categorized by
function or considered collectively was not consistent across
various treatment histories. In fact, TP53 mutations
considered collectively were only associated with OS in patients
who received a certain treatment. TP53 mutations were not
associated with the prognosis in treatment-naïve patients. Such
inconsistencies could be attributed to the following reasons: i)
Not all mutations occurring on hotspot exons (exons 5–8) are
functional; ii) treatment history of treated patients may vary
among patients; iii) the number of patients were significantly
fewer in patients treated with ≥2 lines of treatment; and iv) a
number of studies have reported that TP53 can serve as a
resistance mechanism against the function of EGFR inhibitors
(17,21,44–46).
Therefore, the impact of mutations in TP53 in patients
treated with such inhibitors may be greater compared with patients
treated with other therapies, such as chemotherapy. However,
further examination is required as to why mutations occurring on
exon 8 are associated with unfavorable prognoses. Therefore, exon 8
mutations that potentially serve as prognostic biomarkers require
validation in larger cohorts.
Currently, all TP53 mutations are considered
equally in clinical settings, as well as during the development of
therapeutic strategies, which primarily focuses on the restoration
of the WT activity of TP53 (47). Numerous studies investigating the
prognostic value of TP53 mutations, when all mutations were
considered collectively, identified either no or slight
associations, which was subsequently lost in the multivariate
analysis (22,48). An increasing number of studies have
been categorizing TP53 mutations based on the multiple
biological effects produced by different mutant proteins (15,49).
Notably, the present study strongly followed the aforementioned
approach. Several previous studies categorized TP53
mutations and examined their prognostic value, presenting
conflicting results, partially due to the lack of a unifying
classification system (19,22,50). A
number of studies reported shorter OS in the presence of specific
mutations, including non-disruptive mutations (19), truncated, structural and DNA-binding
mutations (51) or mutations
occurring in certain exons (52,53).
Other studies did not identify an association in patients with lung
cancer (22–24,54).
Some studies have demonstrated that non-disruptive mutations,
allowing the maintenance of functional properties, are associated
with gain-of-function properties (55,56).
Furthermore, mutations occurring on different parts of the gene
have different biological functions such as the AT domain, DBD and
oligomerization domain (25–28). Studies have shown that mutations
occurring in the L2 and L3 domains, providing for DNA contacts, are
associated with poor prognosis (2,28,53). Due
to the discrepancies identified in previous studies, the
development of a clinically relevant unifying classification system
is required. To the best of our knowledge, the present study is the
first that compared the two classification systems commonly used
(based on the position and the type of mutation) in a large
cohort.
Since a significant percentage of patients
exhibiting mutations in EGFR have concurrent TP53
mutations, numerous studies have also assessed the impact of
TP53 mutations on the clinical outcomes of patients with
EGFR mutations treated with EGFR-TKIs (17,21,44–46).
Such studies also yielded conflicting results. A previous study
revealed that the predictive and prognostic power of TP53
status to first-generation EGFR-TKI treatment are more reliable in
patients harboring EGFR exon 19 deletion (19 del) (17). Since TP53 mutations have been
confirmed as a primary resistance mechanism to EGFR-TKI, some
studies reported diminished responses. Canale et al
(17) revealed that TP53 exon
8 mutations, especially in conjunction with EGFR 19
deletion, were associated with a significantly lower disease
control rate. Labbé et al (21) reported a marginally lower response
rate and shorter PFS in patients with concurrent EGFR and
TP53 mutations, where all TP53 mutations were considered
collectively. Collectively, these previous reports and the present
study suggest that the use of a unifying classification system may
be important in clinical settings.
Furthermore, the majority of studies examining the
clinical relevance of TP53 were primarily conducted in
patients with early stage lung cancer and resectable tumors
(20,24,57). To
the best of our knowledge, a few studies investigated patients with
advanced lung cancer and a majority of them included a limited
number of patients (17,19,21,45,54,58). To
the best of our knowledge, the present study is the first study to
investigate, in a large cohort, the clinical relevance of
TP53 mutations in Chinese patients with advanced lung
cancer, who had received previous treatments. Furthermore, to the
best of our knowledge, the present study is also the first one to
investigate the association between TP53 mutations and OS in
patients treated with osimertinib, a TKI inhibitor. It was revealed
that the prognostic power of TP53 mutations only existed in
patients with EGFR 19 del and T790M. In such patients,
non-disruptive mutations were associated with shorter OS. The
prognostic power was not statistically significant in patients
harboring EGFR L858R. A previous study evaluated the impact
of TP53 mutations on the outcomes of patients with
EGFR mutations treated with one course of EGFR-TKI and
revealed similar results (17).
Patients harboring concurrent mutations in the exon 8 of
TP53 and EGFR 19 del were associated with a shorter
PFS and OS. The predictive and prognostic power was much weaker in
subgroups containing patients with other EGFR mutations
(17). One major limitation
associated with the present study is that it only included patients
with classic NSCLC driver mutations. Further examination is
required in order to validate these findings in larger cohorts,
including patients without NSCLC driver mutations.
The results of the present study suggested that not
all TP53 mutations are equal. Mutation in exon 8 can
identify a subgroup of patients with unfavorable prognoses across
diverse treatment group. To the best of our knowledge, the present
study is the first one that compared different TP53 mutation
classification systems in a large cohort of patients with advanced
lung cancer. Furthermore, to the best of our knowledge, the current
study may be the first to reveal that the prognostic potential of
TP53 mutations, in patients treated with osimertinib, only
exists in patients with EGFR 19 del mutation. Further
studies are required to elucidate why TP53 mutations
determined significantly poor prognoses in patients harboring
EGFR 19 del but not in patients presenting EGFR
L858R. The present study may provide novel insights into the
identification of the most optimal treatment strategy.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Wu Jieping
Medical Foundation (grant no. 320.6750.17281).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
RG, LZ, YL and FX conceived and designed the study.
YL, FX, YW, QW, BW, YY, YZ, JL, ZZ, XM and LZ collected the data.
JY performed the statistical analysis of the data. YL, FX, YW, QW,
BW, YY, YZ, HHZ, JY and LZ analyzed and interpreted the data. HHZ,
RG, LZ, YL, FX and LZ wrote the manuscript. All authors approved
the final version of the manuscript and are accountable for all
aspects of the work.
Ethics approval and consent to
participate
All procedures performed involving human subjects
were in accordance with the ethical standards of the Medical Ethics
Committee of Jiangsu Province Hospital (Nanjing, China). All
patients provided written informed consent for participating in the
study.
Patient consent for publication
Not applicable.
Competing interests
HHZ, JZ, LZ, ZZ and JL are employees of Burning
Rock Biotech. The other authors declare that they have no competing
interests.
References
|
1
|
Menendez D, Inga A and Resnick MA: The
expanding universe of p53 targets. Nat Rev Cancer. 9:724–737. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sabapathy K and Lane DP: Therapeutic
targeting of p53: All mutants are equal, but some mutants are more
equal than others. Nat Rev Clin Oncol. 15:13–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lawrence MS, Stojanov P, Mermel CH,
Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander
ES and Getz G: Discovery and saturation analysis of cancer genes
across 21 tumour types. Nature. 505:495–501. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bouaoun L, Sonkin D, Ardin M, Hollstein M,
Byrnes G, Zavadil J and Olivier M: TP53 variations in human
cancers: New lessons from the IARC TP53 database and genomics data.
Hum Mutat. 37:865–876. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bieging KT, Mello SS and Attardi LD:
Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev
Cancer. 14:359–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kruiswijk F, Labuschagne CF and Vousden
KH: p53 in survival, death and metabolic health: A lifeguard with a
licence to kill. Nat Rev Mol Cell Biol. 16:393–405. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vogelstein B, Lane D and Levine AJ:
Surfing the p53 network. Nature. 408:307–310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Merkel O, Taylor N, Prutsch N, Staber PB,
Moriggl R, Turner SD and Kenner L: When the guardian sleeps:
Reactivation of the p53 pathway in cancer. Mutat Res. 773:1–13.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Muller PA and Vousden KH: p53 mutations in
cancer. Nat Cell Biol. 15:2–8. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Freed-Pastor WA and Prives C: Mutant p53:
One name, many proteins. Genes Dev. 26:1268–1286. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Petitjean A, Achatz MI, Borresen-Dale AL,
Hainaut P and Olivier M: TP53 mutations in human cancers:
Functional selection and impact on cancer prognosis and outcomes.
Oncogene. 26:2157–2165. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fong KM, Kida Y, Zimmerman PV, Ikenaga M
and Smith PJ: Loss of heterozygosity frequently affects chromosome
17q in non-small cell lung cancer. Cancer Res. 55:4268–4272.
1995.PubMed/NCBI
|
|
13
|
Olivier M, Eeles R, Hollstein M, Khan MA,
Harris CC and Hainaut P: The IARC TP53 database: New online
mutation analysis and recommendations to users. Hum Mutat.
19:607–614. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Baugh EH, Ke H, Levine AJ, Bonneau RA and
Chan CS: Why are there hotspot mutations in the TP53 gene in human
cancers? Cell Death Differ. 25:154–160. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Poeta ML, Manola J, Goldwasser MA,
Forastiere A, Benoit N, Califano JA, Ridge JA, Goodwin J, Kenady D,
Saunders J, et al: TP53 mutations and survival in squamous-cell
carcinoma of the head and neck. N Engl J Med. 357:2552–2561. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lehmann BD and Pietenpol JA: Targeting
mutant p53 in human tumors. J Clin Oncol. 30:3648–3650. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Canale M, Petracci E, Delmonte A, Chiadini
E, Dazzi C, Papi M, Capelli L, Casanova C, De Luigi N, Mariotti M,
et al: Impact of TP53 mutations on outcome in EGFR-mutated patients
treated with first-line tyrosine kinase inhibitors. Clin Cancer
Res. 23:2195–2202. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rho JK, Choi YJ, Ryoo BY, Na II, Yang SH,
Kim CH and Lee JC: p53 enhances gefitinib-induced growth inhibition
and apoptosis by regulation of Fas in non-small cell lung cancer.
Cancer Res. 67:1163–1169. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Molina-Vila MA, Bertran-Alamillo J, Gascó
A, Mayo-de-las-Casas C, Sánchez-Ronco M, Pujantell-Pastor L,
Bonanno L, Favaretto AG, Cardona AF, Vergnenègre A, et al:
Nondisruptive p53 mutations are associated with shorter survival in
patients with advanced non-small cell lung cancer. Clin Cancer Res.
20:4647–4659. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ma X, Le Teuff G, Lacas B, Tsao MS,
Graziano S, Pignon JP, Douillard JY, Le Chevalier T, Seymour L,
Filipits M, et al: Prognostic and predictive effect of TP53
mutations in patients with non-small cell lung cancer from adjuvant
cisplatin-based therapy randomized trials: A LACE-Bio pooled
analysis. J Thorac Oncol. 11:850–861. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Labbé C, Cabanero M, Korpanty GJ, Tomasini
P, Doherty MK, Mascaux C, Jao K, Pitcher B, Wang R, Pintilie M, et
al: Prognostic and predictive effects of TP53 co-mutation in
patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung
Cancer. 111:23–29. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Scoccianti C, Vesin A, Martel G, Olivier
M, Brambilla E, Timsit JF, Tavecchio L, Brambilla C, Field JK and
Hainaut P; European Early Lung Cancer Consortium, : Prognostic
value of TP53, KRAS and EGFR mutations in nonsmall cell lung
cancer: The EUELC cohort. Eur Respir J. 40:177–184. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee SY, Jeon HS, Hwangbo Y, Jeong JY, Park
JY, Lee EJ, Jin G, Shin KM, Yoo SS, Lee J, et al: The influence of
TP53 mutations on the prognosis of patients with early stage
non-small cell lung cancer may depend on the intratumor
heterogeneity of the mutations. Mol Carcinog. 54:93–101. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma X, Rousseau V, Sun H, Lantuejoul S,
Filipits M, Pirker R, Popper H, Mendiboure J, Vataire AL, Le
Chevalier T, et al: Significance of TP53 mutations as predictive
markers of adjuvant cisplatin-based chemotherapy in completely
resected non-small-cell lung cancer. Mol Oncol. 8:555–564. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lomax ME, Barnes DM, Hupp TR, Picksley SM
and Camplejohn RS: Characterization of p53 oligomerization domain
mutations isolated from Li-Fraumeni and Li-Fraumeni like family
members. Oncogene. 17:643–649. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Phang BH, Othman R, Bougeard G, Chia RH,
Frebourg T, Tang CL, Cheah PY and Sabapathy K: Amino-terminal p53
mutations lead to expression of apoptosis proficient p47 and
prognosticate better survival, but predispose to tumorigenesis.
Proc Natl Acad Sci USA. 112:E6349–E6358. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pavletich NP, Chambers KA and Pabo CO: The
DNA-binding domain of p53 contains the four conserved regions and
the major mutation hot spots. Genes Dev. 7:2556–2564. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dittmer D, Pati S, Zambetti G, Chu S,
Teresky AK, Moore M, Finlay C and Levine AJ: Gain of function
mutations in p53. Nat Genet. 4:42–46. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mao X, Zhang Z, Zheng X, Xie F, Duan F,
Jiang L, Chuai S, Han-Zhang H, Han B and Sun J: Capture-based
targeted ultradeep sequencing in paired tissue and plasma samples
demonstrates differential subclonal ctDNA-releasing capability in
advanced lung cancer. J Thorac Oncol. 12:663–672. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The genome analysis toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Van der Auwera GA, Carneiro MO, Hartl C,
Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen
D, Thibault J, et al: From FastQ data to high confidence variant
calls: The Genome analysis toolkit best practices pipeline. Curr
Protoc Bioinformatics. 43:11.10.1–33. 2013.
|
|
34
|
Koboldt DC, Zhang Q, Larson DE, Shen D,
McLellan MD, Lin L, Miller CA, Mardis ER, Ding L and Wilson RK:
VarScan 2: Somatic mutation and copy number alteration discovery in
cancer by exome sequencing. Genome Res. 22:568–576. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
1000 Genomes Project Consortium, ; Auton
A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini
JL, McCarthy S, McVean GA and Abecasis GR: A global reference for
human genetic variation. Nature. 526:68–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sherry ST, Ward MH, Kholodov M, Baker J,
Phan L, Smigielski EM and Sirotkin K: dbSNP: The NCBI database of
genetic variation. Nucleic Acids Res. 29:308–311. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang K, Li M and Hakonarson H: ANNOVAR:
Functional annotation of genetic variants from high-throughput
sequencing data. Nucleic Acids Res. 38:e1642010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cingolani P, Platts A, Wang le L, Coon M,
Nguyen T, Wang L, Land SJ, Lu X and Ruden DM: A program for
annotating and predicting the effects of single nucleotide
polymorphisms, SnpEff: SNPs in the genome of Drosophila
melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 6:80–92.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Newman AM, Bratman SV, Stehr H, Lee LJ,
Liu CL, Diehn M and Alizadeh AA: FACTERA: A practical method for
the discovery of genomic rearrangements at breakpoint resolution.
Bioinformatics. 30:3390–3393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
RStudio Team, . RStudio: Integrated
Development for R. RStudio, Inc., Boston, MA, 2015. http://www.rstudio.com/
|
|
41
|
Awad MM and Shaw AT: ALK inhibitors in
non-small cell lung cancer: Crizotinib and beyond. Clin Adv Hematol
Oncol. 12:429–439. 2014.PubMed/NCBI
|
|
42
|
Karachaliou N, Sosa AE and Rosell R:
Unraveling the genomic complexity of small cell lung cancer. Transl
Lung Cancer Res. 5:363–366. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Remon J, Steuer CE, Ramalingam SS and
Felip E: Osimertinib and other third-generation EGFR TKI in
EGFR-mutant NSCLC patients. Ann Oncol. 29 (Suppl 1):i20–i27. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Huang S, Benavente S, Armstrong EA, Li C,
Wheeler DL and Harari PM: p53 modulates acquired resistance to EGFR
inhibitors and radiation. Cancer Res. 71:7071–7079. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
VanderLaan PA, Rangachari D, Mockus SM,
Spotlow V, Reddi HV, Malcolm J, Huberman MS, Joseph LJ, Kobayashi
SS and Costa DB: Mutations in TP53, PIK3CA, PTEN and other genes in
EGFR mutated lung cancers: Correlation with clinical outcomes. Lung
Cancer. 106:17–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Iwama E, Sakai K, Azuma K, Harada D,
Nosaki K, Hotta K, Nishio M, Kurata T, Fukuhara T, Akamatsu H, et
al: Exploration of resistance mechanisms for epidermal growth
factor receptor-tyrosine kinase inhibitors based on plasma analysis
by digital polymerase chain reaction and next-generation
sequencing. Cancer Sci. 109:3921–3933. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Blandino G and Di Agostino S: New
therapeutic strategies to treat human cancers expressing mutant p53
proteins. J Exp Clin Cancer Res. 37:302018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kosaka T, Yatabe Y, Onozato R, Kuwano H
and Mitsudomi T: Prognostic implication of EGFR, KRAS, and TP53
gene mutations in a large cohort of Japanese patients with
surgically treated lung adenocarcinoma. J Thorac Oncol. 4:22–29.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Trbusek M, Smardova J, Malcikova J,
Sebejova L, Dobes P, Svitakova M, Vranova V, Mraz M, Francova HS,
Doubek M, et al: Missense mutations located in structural p53
DNA-binding motifs are associated with extremely poor survival in
chronic lymphocytic leukemia. J Clin Oncol. 29:2703–2708. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Szymanowska A, Jassem E, Dziadziuszko R,
Skrzypski M, Kobierska-Gulida G, Holm K, Borg A, Rzyman W, Limon J
and Jassem J: Analysis of prognostic value of TP53 gene mutations
in non-small cell lung cancer. Pneumonol Alergol Pol. 73:264–269.
2005.(In Polish). PubMed/NCBI
|
|
51
|
Ahrendt SA, Hu Y, Buta M, McDermott MP,
Benoit N, Yang SC, Wu L and Sidransky D: p53 mutations and survival
in stage I non-small-cell lung cancer: Results of a prospective
study. J Natl Cancer Inst. 95:961–970. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Huang C, Taki T, Adachi M, Konishi T,
Higashiyama M and Miyake M: Mutations in exon 7 and 8 of p53 as
poor prognostic factors in patients with non-small cell lung
cancer. Oncogene. 16:2469–2477. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Skaug V, Ryberg D, Kure EH, Arab MO,
Stangeland L, Myking AO and Haugen A: p53 mutations in defined
structural and functional domains are related to poor clinical
outcome in non-small cell lung cancer patients. Clin Cancer Res.
6:1031–1037. 2000.PubMed/NCBI
|
|
54
|
Lim EH, Zhang SL, Li JL, Yap WS, Howe TC,
Tan BP, Lee YS, Wong D, Khoo KL, Seto KY, et al: Using whole genome
amplification (WGA) of low-volume biopsies to assess the prognostic
role of EGFR, KRAS, p53, and CMET mutations in advanced-stage
non-small cell lung cancer (NSCLC). J Thorac Oncol. 4:12–21. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Vikhanskaya F, Lee MK, Mazzoletti M,
Broggini M and Sabapathy K: Cancer-derived p53 mutants suppress
p53-target gene expression-potential mechanism for gain of function
of mutant p53. Nucleic Acids Res. 35:2093–2104. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kim MP, Zhang Y and Lozano G: Mutant p53:
Multiple mechanisms define biologic activity in cancer. Front
Oncol. 5:2492015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gu J, Zhou Y, Huang L, Ou W, Wu J, Li S,
Xu J, Feng J and Liu B: TP53 mutation is associated with a poor
clinical outcome for non-small cell lung cancer: Evidence from a
meta-analysis. Mol Clin Oncol. 5:705–713. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Murakami I, Hiyama K, Ishioka S, Yamakido
M, Kasagi F and Yokosaki Y: p53 gene mutations are associated with
shortened survival in patients with advanced non-small cell lung
cancer: An analysis of medically managed patients. Clin Cancer Res.
6:526–530. 2000.PubMed/NCBI
|