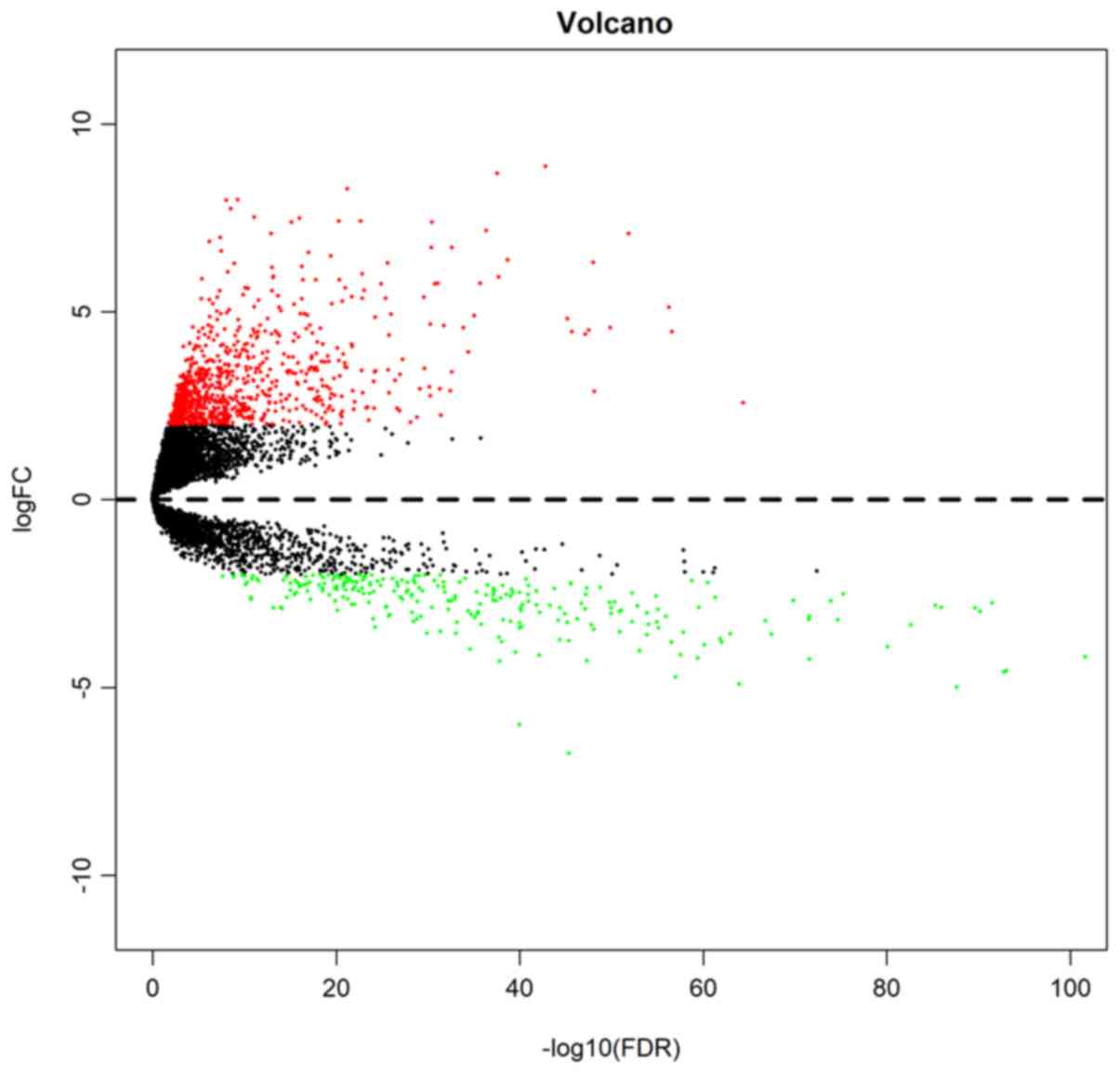
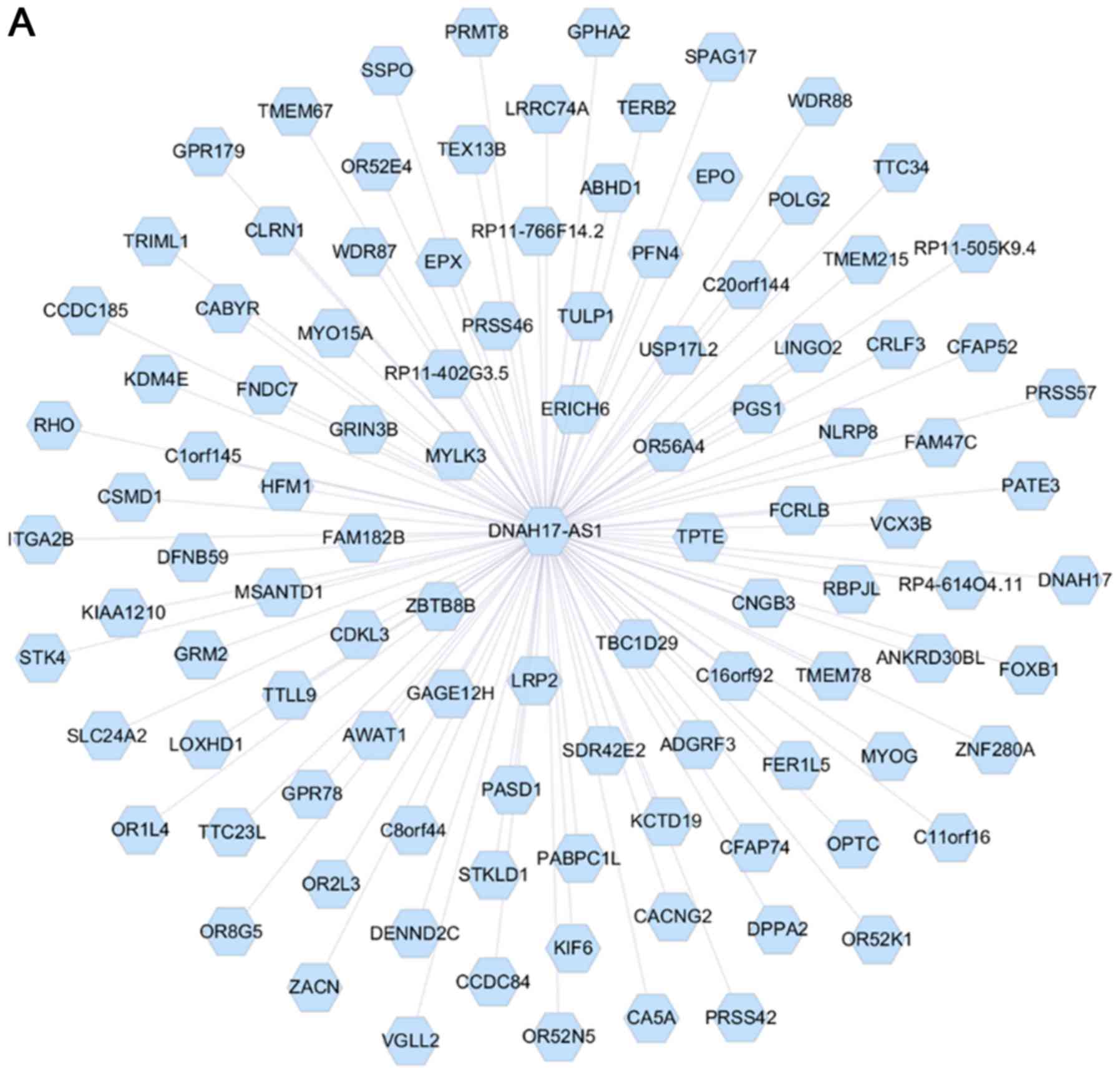
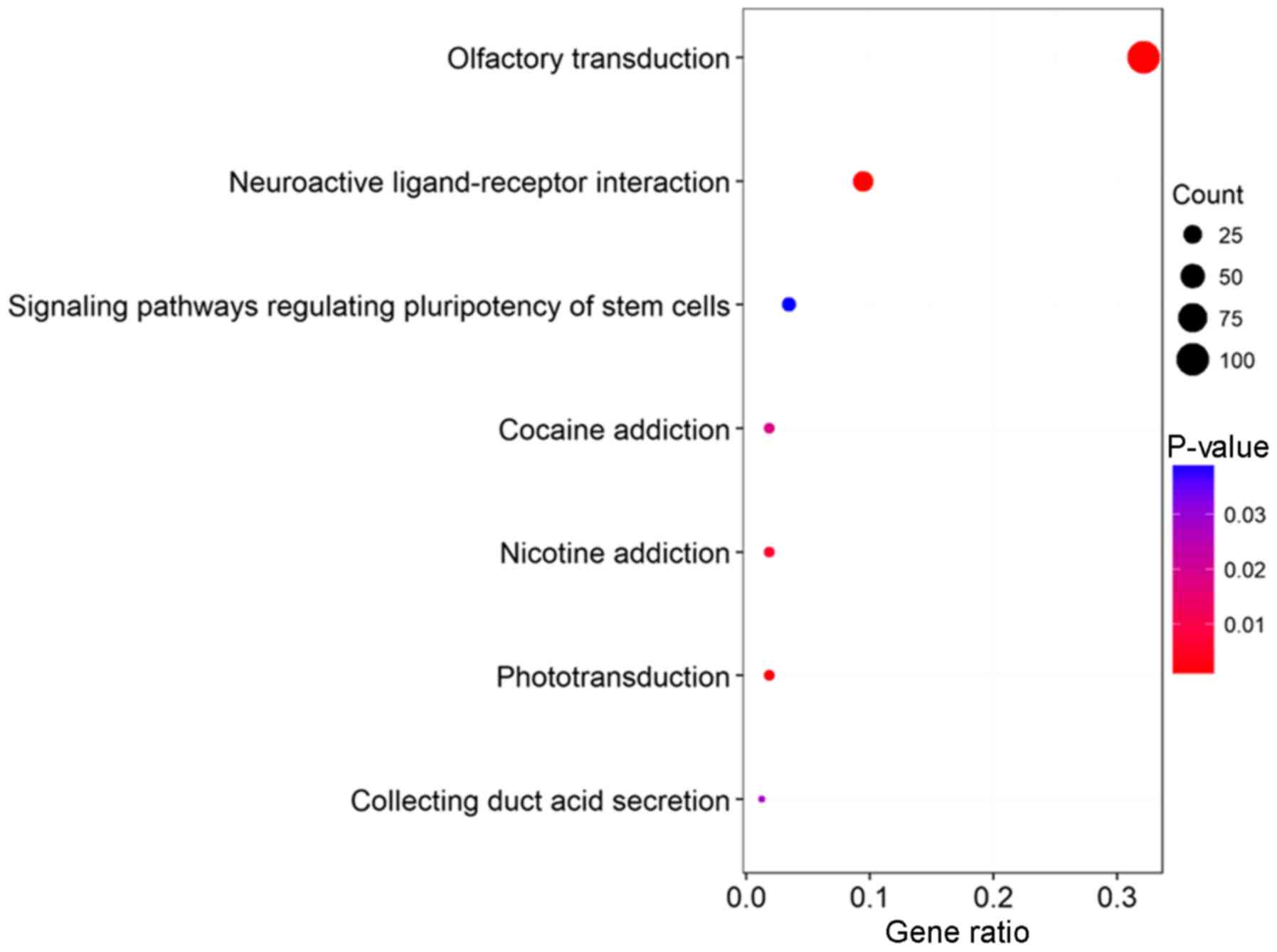
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ghareeb AE, Moawed FSM, Ghareeb DA and
Kandil EI: Potential prophylactic effect of berberine against rat
colon carcinoma induce by 1,2-dimethyl hydrazine. Asian Pac J
Cancer Prev. 19:1685–1690. 2018.PubMed/NCBI
|
|
3
|
Sanoff HK, Sargent DJ, Campbell ME, Morton
RF, Fuchs CS, Ramanathan RK, Williamson SK, Findlay BP, Pitot HC
and Goldberg RM: Five-year data and prognostic factor analysis of
oxaliplatin and irinotecan combinations for advanced colorectal
cancer: N9741. J Clin Oncol. 26:5721–5727. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dong Z, Zheng L, Liu W and Wang C:
Association of mRNA expression of TP53 and the TP53 codon 72
Arg/Pro gene polymorphism with colorectal cancer risk in Asian
population: A bioinformatics analysis and meta-analysis. Cancer
Manag Res. 10:1341–1349. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xue W, Li J, Wang F, Han P, Liu Y and Cui
B: A long non-coding RNA expression signature to predict survival
of patients with colon adenocarcinoma. Oncotarget. 8:101298–101308.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dykes IM and Emanueli C: Transcriptional
and posttranscriptional gene regulation by long non-coding RNA.
Genomics Proteomics Bioinformatics. 15:177–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long non-coding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou M, Wang X, Li J, Hao D, Wang Z, Shi
H, Han L, Zhou H and Sun J: Prioritizing candidate disease-related
long non-coding RNAs by walking on the heterogeneous lncRNA and
disease network. Mol Biosyst. 11:760–769. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lou Y, Jiang H, Cui Z, Wang X, Wang L and
Han Y: Gene microarray analysis of lncRNA and mRNA expression
profiles in patients with high-grade ovarian serous cancer. Int J
Mol Med. 42:91–104. 2018.PubMed/NCBI
|
|
11
|
Jin B, Jin H, Wu HB, Xu JJ and Li B: Long
non-coding RNA SNHG15 promotes CDK14 expression via miR-486 to
accelerate non-small cell lung cancer cells progression and
metastasis. J Cell Physiol. 233:7164–7172. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li S, Zhou J, Wang Z, Wang P, Gao X and
Wang Y: Long noncoding RNA GAS5 suppresses triple negative breast
cancer progression through inhibition of proliferation and invasion
by competitively binding miR-196a-5p. Biomed Pharmacother.
104:451–457. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu Z, Yan Y, Cao S and Chen Y: Long
non-coding RNA SNHG14 contributes to gastric cancer development
through targeting miR-145/SOX9 axis. J Cell Biochem. 119:6905–6913.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fan H, Lv P, Mu T, Zhao X, Liu Y, Feng Y,
Lv J, Liu M and Tang H: lncRNA n335586/miR-924/CKMT1A axis
contributes to cell migration and invasion in hepatocellular
carcinoma cells. Cancer Lett. 429:89–99. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ye XT, Huang H, Huang WP and Hu WL: lncRNA
THOR promotes human renal cell carcinoma cell growth. Biochem
Biophys Res Commun. 501:661–667. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tsai KW, Lo YH, Liu H, Yeh CY, Chen YZ,
Hsu CW, Chen WS and Wang JH: Linc00659, a long noncoding RNA, acts
as novel oncogene in regulating cancer cell growth in colorectal
cancer. Mol Cancer. 17:722018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu Q, Meng WY, Jie Y and Zhao H: lncRNA
MALAT1 induces colon cancer development by regulating
miR-129-5p/HMGB1 axis. J Cell Physiol. 233:6750–6757. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang H, Wang S, Kang YJ, Wang C, Xu Y,
Zhang Y and Jiang Z: Long non-coding RNA SNHG1 predicts a poor
prognosis and promotes colon cancer tumorigenesis. Oncol Rep.
40:261–271. 2018.PubMed/NCBI
|
|
19
|
Xu Y, Zhang X, Hu X, Zhou W, Zhang P,
Zhang J, Yang S and Liu Y: The effects of lncRNA MALAT1 on
proliferation, invasion and migration in colorectal cancer through
regulating SOX9. Mol Med. 24:522018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu M, Chen X, Lin K, Zeng K, Liu X, Pan B,
Xu X, Xu T, Hu X, Sun L, et al: The long noncoding RNA SNHG1
regulates colorectal cancer cell growth through interactions with
EZH2 and miR-154-5p. Mol Cancer. 17:1412018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Edge SB, Byrd DR, Compton CC, Fritz AG
Greene, F and Trotti A: AJCC Cancer Staging Manual. 7th.
Springer-Verlag; New York, NY: 2010
|
|
22
|
Gene Ontology Consortium: The gene
ontology (GO) project in 2006. Nucleic Acids Res. 34:D322–D326.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Altermann E and Klaenhammer TR:
Pathwayvoyager: Pathway mapping using the Kyoto encyclopedia of
genes and genomes (KEGG) database. BMC Genomics. 6:602005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim JH, Sio CA, Park H, Kim H, Shin HD and
Jung K: Undifferentiated embryonal sarcoma of the liver in a child:
A whole exome sequencing analysis. Dig Liver Dis. 49:944–946. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhan H, Jiang J, Sun Q, Ke A, Hu J, Hu Z,
Zhu K, Luo C, Ren N, Fan J, et al: Whole-exome sequencing-based
mutational profiling of hepatitis B virus-related early-stage
hepatocellular carcinoma. Gastroenterol Res Pract 2017.
20293152017.
|
|
26
|
Fan X, Guo H, Dai B, He L, Zhou D and Lin
H: The association between methylation patterns of DNAH17 and
clinicopathological factors in hepatocellular carcinoma. Cancer
Med. 8:337–350. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun L, Jiang C, Xu C, Xue H, Zhou H, Gu L,
Liu Y and Xu Q: Down-regulation of long non-coding RNA
RP11-708H21.4 is associated with poor prognosis of colorectal
cancer and promotes tumorigenesis through regulating AKT/mTOR
pathway. Oncotarget. 8:27929–27942. 2017.PubMed/NCBI
|
|
28
|
Shi D, Zheng H, Zhuo C, Peng J, Li D, Xu
Y, Li X, Cai G and Cai S: Low expression of novel lncRNA
RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal
cancer. Med Oncol. 31:312014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tong W, Yang L, Yu Q, Yao J and He A: A
new tumor suppressor lncRNA RP11-190D6.2 inhibits the
proliferation, migration and invasion of epithelial ovarian cancer
cells. Onco Targets Ther. 10:1227–1235. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang KF, Jin W, Song Y and Fei X: lncRNA
RP11-436H11.5, functioning as a competitive endogenous RNA,
upregulates BCL-W expression by sponging miR-335-5p and promotes
proliferation and invasion in renal cell carcinoma. Mol Cancer.
16:1662017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hasenoehrl C, Feuersinger D, Sturm EM,
Bärnthaler T, Heitzer E, Graf R, Grill M, Pichler M, Beck S,
Butcher L, et al: G protein-coupled receptor GPR55 promotes
colorectal cancer and has opposing effects to cannabinoid receptor
1. Int J Cancer. 142:121–132. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kargl J, Andersen L, Hasenöhrl C,
Feuersinger D, Stančić A, Fauland A, Magnes C, El-Heliebi A, Lax S,
Uranitsch S, et al: GPR55 promotes migration and adhesion of colon
cancer cells indicating a role in metastasis. Br J Pharmacol.
173:142–154. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bai B, Chen X, Zhang R, Wang X, Jiang Y,
Li D, Wang Z and Chen J: Dual-agonist occupancy of orexin receptor
1 and cholecystokinin A receptor heterodimers decreases
G-protein-dependent signaling and migration in the human colon
cancer cell line HT-29. Biochim Biophys Acta Mol Cell Res 1864.
1153–1164. 2017. View Article : Google Scholar
|
|
34
|
Bardhan K, Paschall AV, Yang D, Chen MR,
Simon PS, Bhutia YD, Martin PM, Thangaraju M, Browning DD,
Ganapathy V, et al: IFNγ Induces DNA methylation-silenced GPR109A
expression via pSTAT1/p300 and H3K18 acetylation in colon cancer.
Cancer Immunol Res. 3:795–805. 2015. View Article : Google Scholar : PubMed/NCBI
|