Introduction
Prostate adenocarcinoma (PRAD) is one of the most
common causes of cancer-associated mortality worldwide (1,2). PRAD is
the most frequent prostate cancer histological subtype worldwide,
and is derived from basal cells differentiating into glandular
cells (3). The annual morbidity rate
of PRAD has grown steadily, and the incidence of this cancer has
increased by 14% over the last two decades worldwide (4,5).
Approximately 30% of men with the disease develop clinical
recurrence, and this highly aggressive form of PRAD can lead to
mortality (6). One method of curing
PRAD is gene therapy (7). Thus, the
identification and analysis of survival-associated biomarkers
provides key opportunities for improving the prognosis of
patients.
Long non-coding RNAs (lncRNAs) are defined as
non-protein coding transcripts >200 nucleotides in length
(8), and they play an important role
in various types of cancer (9), such
as colon cancer, breast cancer and PRAD (10,11).
Prostate cancer antigen 3, a well-characterized lncRNA, has been
approved by the US Food and Drug Administration for clinical
decisions regarding repeat biopsies for prostate cancer (12). lncRNAs, and their associated genes,
can be negatively regulated by microRNAs (miRNAs) (13–15).
Accumulating evidence has suggested that lncRNAs and protein coding
genes could serve as competitive endogenous RNAs (ceRNAs), which
exert their decoy activity by recruiting miRNA molecules via
base-pairing with miRNA-recognition elements (MREs), which
subsequently compete with common miRNAs, thus contributing to tumor
development, progression and metastasis (16,17). For
example, in multiple myeloma, lncRNA metastasis-associated lung
adenocarcinoma transcript 1 acts as an oncogene by sponging
miR-509-5p to modulate forkhead box P1 expression (18). Based on the ceRNA mechanism, miRNAs
and ceRNAs could influence the expression of one another (19). For instance, phosphatase and tensin
homolog pseudogene 1 functions as a ceRNA in order to modulate
levels of PTEN by decoying miRNA (miR)-106b and miR-93 in gastric
cancer (20). Recently, ceRNA-based
computational methods have been used in cancer-associated studies
(21,22). Wu et al (23) investigated the miR-133b-mediated
lncRNA-mRNA ceRNA network and laid the foundation for future
investigation into the role of lncRNAs in colorectal cancer.
Another study demonstrated that indirect interactions among
networks of ceRNAs may aid in improving responses to cancer therapy
and the development of new therapeutic interventions (24). However, the role of ceRNAs in PRAD
remains to be fully investigated.
The aim of the present study was to provide further
insights into the current understanding of the potential
interactions of ceRNA triplets, and provide potential prognostic
markers for PRAD.
Materials and methods
Expression of genes, lncRNAs and
miRNAs
First, the RNA-sequencing version 2 datasets for
PRAD were downloaded from The Cancer Genome Atlas (TCGA) database
(http://tcga-data.nci.nih.gov/) (25,26). Raw
read counts for each exon were derived from exon quantification
files provided by TCGA level three dataset (the calculated
expression value of a particular composite exon of a gene, per
sample) (27). Then, the reads per
kilobases per million reads (RPKM) value of one gene or lncRNA
could be obtained using the following formula:
RPKM=ECx109/SCxl, in which EC represents mapped read
counts on all exons of one gene or lncRNA, SC represents mapped
read counts on all exons of a sample, and 1 represents the total
length of all exons on a gene or lncRNA. The exon structures of
genes and lncRNAs were downloaded from GENCODE (version 14;
http://www.gencodegenes.org/). The
expression dataset of miRNA (level three) was also obtained from
TCGA database (25). Finally, the
matched RPKMs of genes, lncRNAs and miRNA expression profiles were
extracted from 494 tumor samples and 52 normal samples (25).
Construction of the ceRNA network of
PRAD
The miRNA-target gene interactions were downloaded
from the mirTarBase (28) and
TarBase (29) databases.
Experimental and computational methods were used to predict
miRNA-lncRNA interactions. First, miRanda (30), TargetScan (31), PITA (32) and RNAhybrid (33) were used to predict candidate
miRNA-lncRNA interactions with default parameters. Experimental
interactions in starBase (34) and
DIANA-LncBase (35) were used to
filter candidate miRNA-lncRNA interactions, which provided
high-throughput and photoactivatable ribonucleoside-enhanced
cross-linking and immunoprecipiation experimental data. Next, the
correlations between miRNA and target coding genes/lncRNAs were
calculated using Pearson's correlation coefficient (PCC) using R
software. Due to the negative regulatory relationship between
miRNAs and their targets (genes/lncRNAs), only interactions with a
PCC <-0.1 and P<0.01 were used to construct the PRAD ceRNA
network.
Comparison of PRAD-risk lncRNAs and
non-disease lncRNAs
PRAD-risk lncRNAs were downloaded from LncRNADisease
(36), which stores manually
corrected associations between diseases and lncRNAs. To compare
PRAD-risk lncRNAs and non-disease lncRNAs, the degree, betweenness
centrality, closeness centrality and shortest path were analyzed
for PRAD-risk lncRNAs and non-disease lncRNAs in the ceRNA
network.
Identifying differentially expressed
triplets in PRAD
First, the fold-change method was applied to
identify differentially expressed miRNAs, mRNAs and lncRNAs between
PRAD samples and normal samples with their corresponding RPKM
profile. A molecule was considered to be differentially expressed
with |log2fold change| >1. Subsequently, differentially
expressed triplets (lncRNA-miRNA-mRNA) were extracted from the PRAD
ceRNA network; each element of these triplets was differentially
expressed in PRAD.
Identifying survival-associated
triplets in PRAD
All PRAD-survival-associated triplets were extracted
from the identified candidates, and a weighted expression (WE)
score was calculated for each differentially expressed triplet for
identification. For a triplet i, explnci,
expmiri and expgenei were defined as
representing the normalized expression value of lncRNA, miRNA and
gene in this triplet, respectively. wlnc and
wmir represent the weighted score of lncRNA and miRNA
expression, respectively. The WE scores of triplets (i) were
calculated using the following formula:
WEi=wlnc × explnci +
expgenei + wmir × expmiri. The
expression values of lncRNAs were normalized with the following
formula: explnc=[x-min (x)]/[max(x)-min(x)], in which
min(x) and max(x) represent the minimum and maximum expression
values of the lncRNA, respectively. Using the same method,
expmiri and expgenei could be obtained.
wlnc was calculated using the mean expression of all
genes divided by the mean expression of all lncRNAs. Furthermore,
wmir could be calculated by the mean expression of all
genes divided by the mean expression of all miRNAs. For
differentially expressed triplets, the median WE score was used as
a threshold to classify all the PRAD tumor samples into high-risk
(WE score >31.69) and low-risk groups (WE score <31.69). A
Kaplan-Meier analysis, a common measurement of the fraction of
patients living for a certain amount of time between two groups
(37), was performed to test for
significance between the two groups. The log-rank test was used to
evaluate the significance of differences in survival time. Triplets
with P<0.05 were identified as significant prognostic
biomarkers.
Topological measurement
In a given graph (G), G=(V, E), in which V
represents a set of nodes, and E represents a set of edges. The
degree (D) is defined as the number of edges that connect to a
node. If it is assumed that there are k edges linked to node v,
then the degree of node v could be described as: D(v)=k.
Betweenness centrality measures the centrality of
the node in a network and is defined as the number of shortest
paths from each node to all others that pass through this node; it
reflects the amount of control that a node exerts over the
interactions of other nodes in the network. The betweenness
centrality (BC) of node V is described as: BC(v)=[Σs≠v≠t
(σst(v)/σst)], where σst
represents the number of shortest paths from node s to node t, and
σst(v) represents the number of those paths that pass
through node v.
The closeness centrality is how close a node is to
other nodes in the network and is defined as the average mean path
from this node to all other nodes. The closeness centrality (C) of
node v is defined as:
C(v)=1∑und(u,v)
Where d(u,v) is the shortest distance between node u
and node v, and n is the number of nodes of the network.
These topological measures were analyzed and
visualized using Cytoscape (version 3.2.1) (38).
Results
ceRNA network for PRAD
To construct a PRAD ceRNA network, interactions
between miRNAs and target genes were downloaded from TarBase and
mirTarBase with experimentally supported datasets, and miRNA-lncRNA
interactions were predicted using a computational program and
filtered using experimental data. For miRNA-lncRNA interactions,
interactions predicted by more than two programs (from either
TargetScan, miRanda, PITA or RNAhybrid) were kept as candidates,
and then filtered by experimental interactions in starBase and
DIANA-LncBase. Following de-redundancy, 43,497 validated
miRNA-target pairs and 314,729 miRNA-lncRNA interactions were kept
for further analysis. Next, a PCC <-0.1 and P<0.01 were used
as a cut-off to filter negatively regulated associations between
the miRNAs-genes and lncRNAs. Finally, a ceRNA network for PRAD was
constructed, including 2,159 interactions (516 miRNA-gen
interactions and 1,643 miRNA-lncRNA interactions) between 210
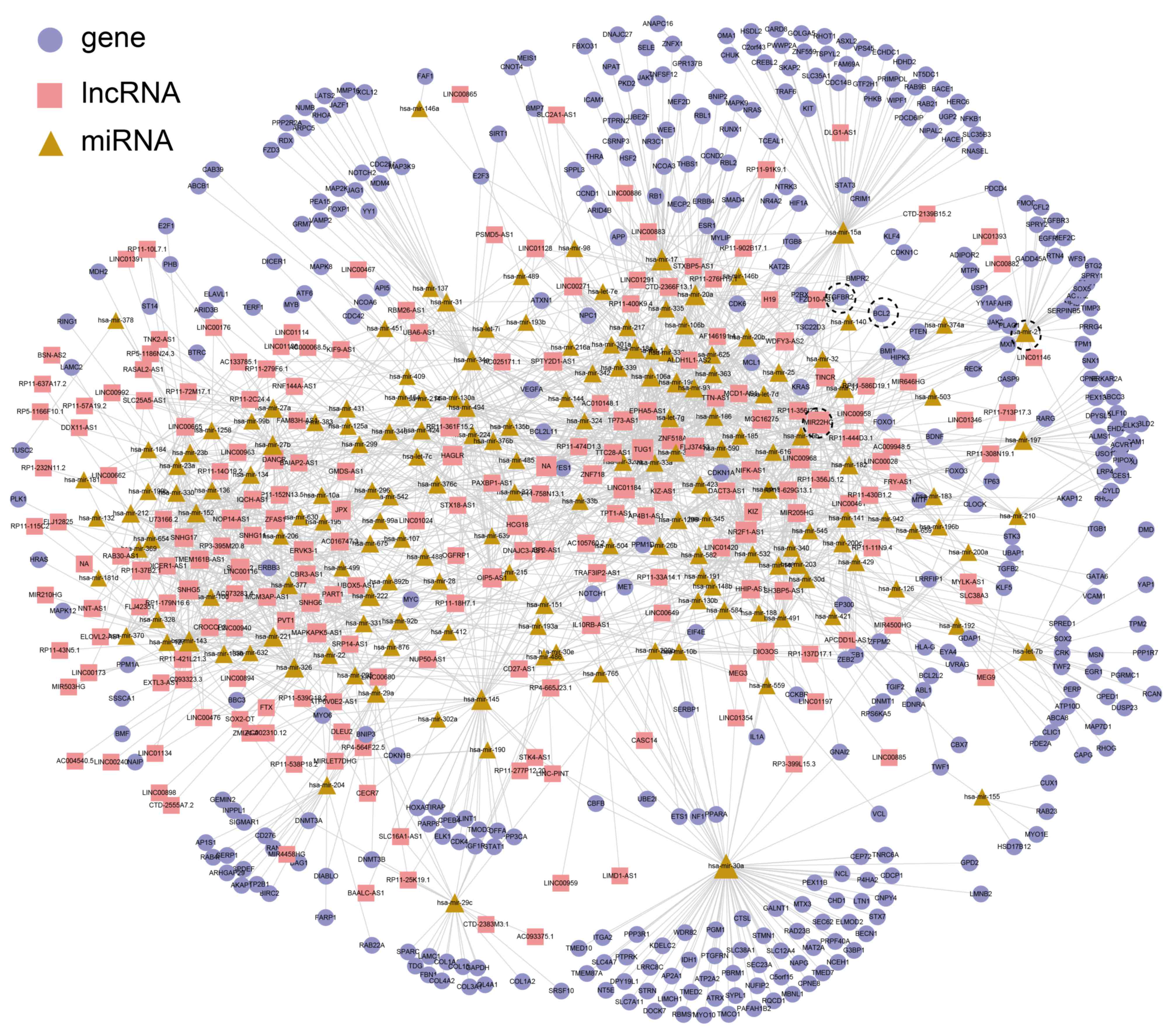
lncRNAs, 169 miRNAs and 391 genes (Fig.
1). To dissect the topological characteristics of the ceRNA
network for PRAD, the degree of node distribution was analyzed
within the network. The ceRNA network for PRAD followed the
power-law distribution with an R2 of 0.893 and a slope
of −1.398 (Fig. 2A). When comparing
the three types of node, it was revealed that the degrees of the
miRNA nodes were higher than those of the genes and lncRNAs,
indicating that miRNAs accounted for the major proportion, whereas
genes and lncRNAs accounted for only a small proportion of the
ceRNA network (Fig. 2B and C).
Comparison of PRAD-risk lncRNAs and
non-disease lncRNAs
PRAD-risk lncRNAs were obtained from LncRNA Disease.
There were six PRAD-risk lncRNAs in the PRAD ceRNA network [H19,
LINC00963, maternally expressed gene 3, differentiation
antagonizing non-protein coding RNA, PVT1 and taurine upregulated
gene 1 (TUG1)]. In order to compare PRAD-risk lncRNAs with
non-disease lncRNA, the topological characteristics of degree,
betweenness centrality, closeness centrality and shortest path were
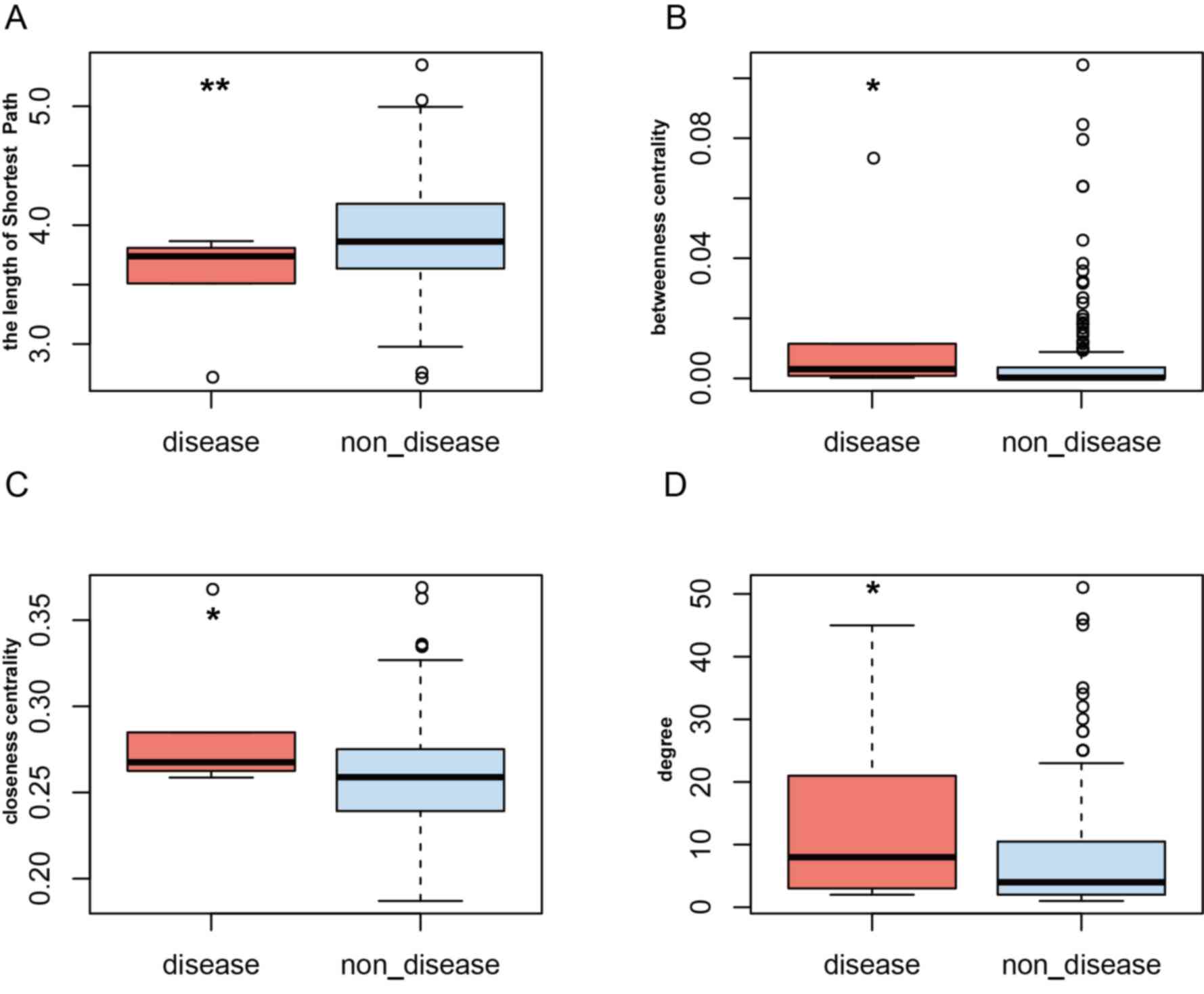
calculated within the PRAD ceRNA network. It was revealed that
PRAD-risk lncRNAs had a higher degree, closeness and betweenness
centrality, but a lower shortest path length (Fig. 3). For example, the degree of TUG1,
which functions as a tumor suppressor by regulating PTEN expression
in prostate cancer (39), ranked
fourth among all of the lncRNAs (degree=45; Fig. 2A).
Survival-associated triplets in
PRAD
To identify survival-associated triplets,
differentially expressed triplets (lncRNA-miRNA-mRNA) were first
extracted from the PRAD ceRNA network, meaning that each element of
the triplet was differentially expressed. There were 159
differentially expressed triplets. WE scores were calculated for
each differentially expressed triplet. All patients with PRAD were
classified into a high-risk group or a low-risk group according to
the median WE score for each differentially expressed triplet. The
Kaplan-Meier analysis was then performed to test the significance
of the two groups. Finally, a total of 42 survival-associated
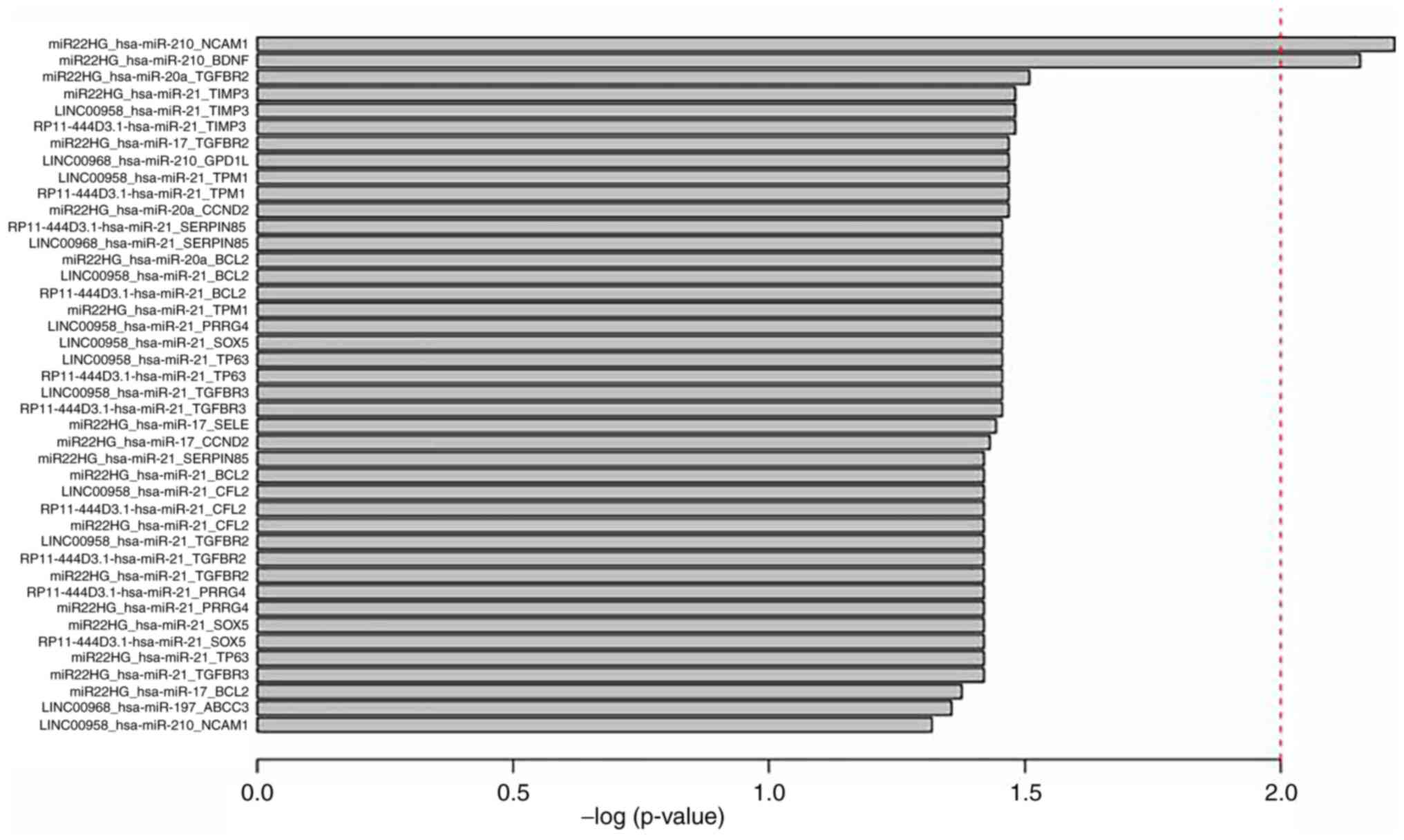
triplets were identified (P<0.05; Table I and Fig.
4).
 | Table I.A total of 42 survival-associated
triplets. |
Table I.
A total of 42 survival-associated
triplets.
| Triplets | P-value |
|---|
|
MIR22HG_hsa-mir-210_NCAM1 | 0.006 |
|
MIR22HG_hsa-mir-210_BDNF | 0.007 |
|
MIR22HG_hsa-mir-20a_TGFBR2 | 0.031 |
|
RP11-444D3.1_hsa-mir-21_TIMP3 | 0.033 |
|
LINC00958_hsa-mir-21_TIMP3 | 0.033 |
|
MIR22HG_hsa-mir-21_TIMP3 | 0.033 |
|
MIR22HG_hsa-mir-20a_CCND2 | 0.034 |
|
RP11-444D3.1_hsa-mir-21_TPM1 | 0.034 |
|
LINC00958_hsa-mir-21_TPM1 | 0.034 |
|
LINC00958_hsa-mir-210_GPD1L | 0.034 |
|
MIR22HG_hsa-mir-17_TGFBR2 | 0.034 |
|
RP11-444D3.1_hsa-mir-21_TGFBR3 | 0.035 |
|
LINC00958_hsa-mir-21_TGFBR3 | 0.035 |
|
RP11-444D3.1_hsa-mir-21_TP63 | 0.035 |
|
LINC00958_hsa-mir-21_TP63 | 0.035 |
|
LINC00958_hsa-mir-21_SOX5 | 0.035 |
|
LINC00958_hsa-mir-21_PRRG4 | 0.035 |
|
MIR22HG_hsa-mir-21_TPM1 | 0.035 |
|
RP11-444D3.1_hsa-mir-21_BCL2 | 0.035 |
|
LINC00958_hsa-mir-21_BCL2 | 0.035 |
|
MIR22HG_hsa-mir-20a_BCL2 | 0.035 |
|
LINC00958_hsa-mir-21_SERPINB5 | 0.035 |
|
RP11-444D3.1_hsa-mir-21_SERPINB5 | 0.035 |
|
MIR22HG_hsa-mir-17_SELE | 0.036 |
|
MIR22HG_hsa-mir-17_CCND2 | 0.037 |
|
MIR22HG_hsa-mir-21_TGFBR3 | 0.038 |
|
MIR22HG_hsa-mir-21_TP63 | 0.038 |
|
RP11-444D3.1_hsa-mir-21_SOX5 | 0.038 |
|
MIR22HG_hsa-mir-21_SOX5 | 0.038 |
|
MIR22HG_hsa-mir-21_PRRG4 | 0.038 |
|
RP11-444D3.1_hsa-mir-21_PRRG4 | 0.038 |
|
MIR22HG_hsa-mir-21_TGFBR2 | 0.038 |
|
RP11-444D3.1_hsa-mir-21_TGFBR2 | 0.038 |
|
LINC00958_hsa-mir-21_TGFBR2 | 0.038 |
|
MIR22HG_hsa-mir-21_CFL2 | 0.038 |
|
RP11-444D3.1_hsa-mir-21_CFL2 | 0.038 |
|
LINC00958_hsa-mir-21_CFL2 | 0.038 |
|
MIR22HG_hsa-mir-21_BCL2 | 0.038 |
|
MIR22HG_hsa-mir-21_SERPINB5 | 0.038 |
|
MIR22HG_hsa-mir-17_BCL2 | 0.042 |
|
LINC00968_hsa-mir-197_ABCC3 | 0.044 |
|
LINC00958_hsa-mir-210_NCAM1 | 0.048 |
Of these triplets, it was revealed that some genes,
miRNAs or lncRNAs were involved in a number of the triplets
identified. These triplets were then extracted and a
survival-associated sub-network was constructed. Notably, these 42
survival-associated triplets were composed of only 25 nodes (5
miRNAs, 4 lncRNAs and 16 genes) and 32 edges (Fig. 5A). The miRNA with the highest degree
was miR-21. The lncRNAs and genes with most connections were
MIR22HG, and transforming growth factor-β receptor 2 (TGFBR2) and
BCL2, respectively. Thus, two triplets were identified
(MIR22HG_hsa-mir-21_TGFBR2 and MIR22HG_hsa-mir-21_BCL2). The
survival curves of these triplets are presented in Fig. 5B and C, respectively. The red curve
represents samples with a higher WE score and the blue curve
represents samples with a lower WE score.
Discussion
The ceRNA hypothesis has been reported to represent
a novel post-transcriptional layer of gene regulation operating via
miRNA competition (16). With the
crosstalk of ceRNAs, it has been demonstrated that miRNAs, and
their ceRNA targets, can connect directly or indirectly to form a
complex ceRNA network (40). In the
present study, a PRAD-specific ceRNA network was constructed by
integrating experimentally validated and computationally predicted
miRNA-targeted gene/lncRNA interactions, as well as the negative
expression correlations between miRNA-target genes and lncRNAs. The
ceRNA network for PRAD contained 2,159 interactions (516 miRNA-gen
interactions and 1,643 miRNA-lncRNA interactions) between 210
lncRNAs, 169 miRNAs and 391 genes. The ceRNA network followed the
power-law distribution with an R2 of 0.893 and a slope
of −1.398 (Fig. 2A). lncRNAs with
the largest degree value indicates it has an important role in
PRAD. In the present study, the average degree, shortest path,
betweenness centrality and closeness centrality between PRAD-risk
lncRNAs and non-disease lncRNAs were compared. PRAD-risk lncRNAs
demonstrated a higher degree, closeness centrality and betweenness
centrality, but a lower shortest path length, suggesting that they
were more important in communication and the diffusion of
information than non-disease nodes in the ceRNA network. Next,
differentially expressed lncRNA-miRNA-gene triplets were identified
as candidates and PRAD survival-associated triplets were extracted.
Finally, 42 PRAD-survival-associated triplets were obtained.
Notably, these triplets constructed a compact network composed of
only 25 nodes and 32 edges, indicating that some nodes were
included in a number of triplets. It was demonstrated that there
were only four lncRNAs (LINC00968, MIR22HG, RP11-444D3.1 and
LINC00958) within this subnetwork. To the best of our knowledge,
there have been no previous reports of these lncRNAs and their
association with PRAD, although they have been previously
associated with different types of cancer. LINC00968 and MIR22HG
were revealed to be differentially expressed in lung squamous cell
carcinoma (41,42), while LINC00958 was previously
identified as a candidate oncogene in bladder cancer (43).
The miRNA with the highest degree was miR-21, which
may target and inhibit the tumor suppressor gene PTEN to promote
the proliferation and invasion of prostate cancer cells (44). The genes with the highest degrees
were TGFBR2 and BCL2, which are apoptosis regulators associated
with many different types of cancer. The TGF-β family serves a
fundamental role in a number of different cellular functions in a
developmental, context-dependent and cell type-specific manner
(e.g., cell migration, survival, proliferation and differentiation)
(45). TGFBR2 has previously been
indicated to act as a tumor suppressor gene (46,47).
Finally, two triplets (MIR22HG_hsa-mir-21_TGFBR2 and
MIR22HG_hsa-mir-21_BCL2) were identified in the present study that
were not only associated with PRAD survival but also had the
highest average degree in the sub-network (Fig. 5A). Notably, it was revealed that
although these two triplets were significant as a whole, the nodes
in these triplets were not significantly associated with PRAD
survival alone. It has been reported that upregulation of miR-21
may serve as an independent predictor of progress-free survival in
patients with advanced prostate cancer (48,49). In
addition, miR-21 could exert its oncogenic effects in prostate
tumors by downregulating TGFBR2, thus inhibiting the tumor
suppressive activity of the TGF-β pathway (50). The triplet results from the present
study complement these previous studies, suggesting that miR-21 may
be outcompeted by ceRNAs (MIR22HG, TGFBR2 and BCL2), and play an
important role in PRAD. miRNAs have been proposed as promising
anticancer therapeutic targets (51). As miRNAs are located in the center of
the ceRNA network, it is reasonable to envision the potential of
these cancer-associated ceRNAs as therapeutic targets.
The success of the present study can be attributed
to two aspects: First, the interactions used to construct the
PRAD-specific ceRNA network were supported by both experimental and
computational approaches. Furthermore, the negative expression
regulatory mechanisms between miRNAs and their targets
(lncRNAs/genes) in PRAD were also considered. This provided more
accuracy in characterizing the ceRNA regulatory associations with
PRAD. Secondly, it is more reasonable to identify
survival-associated triplets by integrating information from the
lncRNAs, genes, miRNAs and topological features, which could
therefore help us to understand the ceRNA interactions and the role
of lncRNAs in PRAD.
In summary, survival-associated ceRNA triplets
involved in PRAD were identified in the present study by
constructing a PRAD-specific ceRNA network. These ceRNA triplets
may serve as potential therapeutic targets and prognostic
biomarkers for PRAD, and the results provide important insights
into the understanding of the potential interactions of ceRNA in
PRAD.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YH designed the study. FL and HL collated the data,
designed and developed the database, performed data analyses, and
produced the initial draft of the manuscript. FL contributed to
revising of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang X, Yuan T, Liang M, Du M, Xia S,
Dittmar R, Wang D, See W, Costello BA, Quevedo F, et al: Exosomal
miR-1290 and miR-375 as prognostic markers in castration-resistant
prostate cancer. Eur Urol. 67:33–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kar S, Sengupta D, Deb M, Pradhan N and
Patra SK: SOX2 function and Hedgehog signaling pathway are
co-conspirators in promoting androgen independent prostate cancer.
Biochim Biophys Acta Mol Basis Dis. 1863:253–265. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Parnes HL, House MG and Tangrea JA:
Prostate cancer prevention: Strategies for agent development. Curr
Opin Oncol. 25:242–251. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun X, Yang Z, Zhang Y, He J, Wang F, Su
P, Han J, Song Z and Fei Y: Prognostic implications of tissue and
serum levels of microRNA-128 in human prostate cancer. Int J Clin
Exp Pathol. 8:8394–8401. 2015.PubMed/NCBI
|
|
6
|
Das DK, Osborne JR, Lin HY, Park JY and
Ogunwobi OO: miR-1207-3p is a novel prognostic biomarker of
prostate cancer. Transl Oncol. 9:236–241. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ling CQ, Wang LN, Wang Y, Zhang YH, Yin
ZF, Wang M and Ling C: The roles of traditional Chinese medicine in
gene therapy. J Integr Med. 12:67–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Perkel JM: Visiting ‘noncodarnia’.
Biotechniques. 54:301, 303–304. 2013. View Article : Google Scholar
|
|
9
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu K, Yao H, Wen Y, Zhao H, Zhou N, Lei S
and Xiong L: Functional role of a long non-coding RNA
LIFR-AS1/miR-29a/TNFAIP3 axis in colorectal cancer resistance to
pohotodynamictherapy. Biochim Biophys Acta Mol Basis Dis.
1864:2871–2880. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bolha L, Ravnik-Glavac M and Glavac D:
Long noncoding RNAs as biomarkers in cancer. Dis Markers.
2017:72439682017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Groskopf J, Aubin SM, Deras IL, Blase A,
Bodrug S, Clark C, Brentano S, Mathis J, Pham J, Meyer T, et al:
APTIMA PCA3 molecular urine test: Development of a method to aid in
the diagnosis of prostate cancer. Clin Chem. 52:1089–1095. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yoon JH, Abdelmohsen K and Gorospe M:
Functional interactions among microRNAs and long noncoding RNAs.
Semin Cell Dev Biol. 34:9–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li SQ, Li F, Xiao Y, Wang CM, Tuo L, Hu J,
Yang XB, Wang JS, Shi WH, Li X and Cao XF: Comparison of long
noncoding RNAs, microRNAs and messenger RNAs involved in initiation
and progression of esophageal squamous cell carcinoma. Mol Med Rep.
10:652–662. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thum T and Condorelli G: Long noncoding
RNAs and microRNAs in cardiovascular pathophysiology. Circ Res.
116:751–762. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tay Y, Karreth FA and Pandolfi PP:
Aberrant ceRNA activity drives lung cancer. Cell Res. 24:259–260.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gu Y, Xiao X and Yang S: LncRNA MALAT1
acts as an oncogene in multiple myeloma through sponging miR-509-5p
to modulate FOXP1 expression. Oncotarget. 8:101984–101993. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tripathi V, Shen Z, Chakraborty A, Giri S,
Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A and
Prasanth KV: Long noncoding RNA MALAT1 controls cell cycle
progression by regulating the expression of oncogenic transcription
factor B-MYB. PLoS Genet. 9:e10033682013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang R, Guo Y, Ma Z, Ma G, Xue Q, Li F
and Liu L: Long non-coding RNA PTENP1 functions as a ceRNA to
modulate PTEN level by decoying miR-106b and miR-93 in gastric
cancer. Oncotarget. 8:26079–26089. 2017.PubMed/NCBI
|
|
21
|
Samir N, Matboli M, El-Tayeb H, El-Tawdi
A, Hassan MK, Waly A, El-Akkad HAE, Ramadan MG, Al-Belkini TN,
El-Khamisy S and El-Asmar F: Competing endogenous RNA network
crosstalk reveals novel molecular markers in colorectal cancer. J
Cell Biochem. 8:6869–6881. 2018. View Article : Google Scholar
|
|
22
|
An Y, Furber K and Ji S: Pseudogenes
regulate parental gene expression via ceRNA network. J Cell Mol
Med. 21:185–192. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu H, Wu R, Chen M, Li D, Dai J, Zhang Y,
Gao K, Yu J, Hu G, Guo Y, et al: Comprehensive analysis of
differentially expressed profiles of lncRNAs and construction of
miR-133b mediated ceRNA network in colorectal cancer. Oncotarget.
8:21095–21105. 2017.PubMed/NCBI
|
|
24
|
Giza DE, Vasilescu C and Calin GA:
MicroRNAs and ceRNAs: Therapeutic implications of RNA networks.
Expert Opin Biol Ther. 14:1285–1293. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cancer Genome Atlas Research Network, .
The molecular taxonomy of primary prostate cancer. Cell.
163:1011–1025. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xue D, Lu H, Xu HY, Zhou CX and He XZ:
Long noncoding RNA MALAT1 enhances the docetaxel resistance of
prostate cancer cells via miR-145-5p-mediated regulation of AKAP12.
J Cell Mol Med. 22:3223–3237. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Z, Dai J and Shen H: Systematic
analysis reveals long noncoding RNAs regulating neighboring
transcription factors in human cancers. Biochim Biophys Acta Mol
Basis Dis. 9:2785–2792. 2018. View Article : Google Scholar
|
|
28
|
Chou CH, Chang NW, Shrestha S, Hsu SD, Lin
YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al: miRTarBase
2016: Updates to the experimentally validated miRNA-target
interactions database. Nucleic Acids Res. 44:D239–D247. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vlachos IS, Paraskevopoulou MD, Karagkouni
D, Georgakilas G, Vergoulis T, Kanellos I, Anastasopoulos IL,
Maniou S, Karathanou K and Kalfakakou D: DIANA-TarBase v7.0:
Indexing more than half a million experimentally supported miRNA:
mRNA interactions. Nucleic Acids Res. 43((Database Issue)):
D153–D159. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11:R902010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rehmsmeier M, Steffen P, Hochsmann M and
Giegerich R: Fast and effective prediction of microRNA/target
duplexes. RNA. 10:1507–1517. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42((Database Issue)): D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM and Hatzigeorgiou
AG: DIANA-LncBase: Experimentally verified and computationally
predicted microRNA targets on long non-coding RNAs. Nucleic Acids
Res. 41((Database Issue)): D239–D245. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen
X, Zhang Q, Yan G and Cui Q: LncRNADisease: A database for
long-non-coding RNA-associated diseases. Nucleic Acids Res.
41((Database Issue)): D983–D986. 2013.PubMed/NCBI
|
|
37
|
Rich JT, Neely JG, Paniello RC, Voelker
CC, Nussenbaum B and Wang EW: A practical guide to understanding
Kaplan-Meier curves. Otolaryngol Head Neck Surg. 143:331–336. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Du Z, Sun T, Hacisuleyman E, Fei T, Wang
X, Brown M, Rinn JL, Lee MG, Chen Y, Kantoff PW and Liu XS:
Integrative analyses reveal a long noncoding RNA-mediated sponge
regulatory network in prostate cancer. Nat Commun. 7:109822016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Karreth FA and Pandolfi PP: ceRNA
cross-talk in cancer: When ce-bling rivalries go awry. Cancer
Discov. 3:1113–1121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen WJ, Tang RX, He RQ, Li DY, Liang L,
Zeng JH, Hu XH, Ma J, Li SK and Chen G: Clinical roles of the
aberrantly expressed lncRNAs in lung squamous cell carcinoma: A
study based on RNA-sequencing and microarray data mining.
Oncotarget. 8:61282–61304. 2017.PubMed/NCBI
|
|
42
|
Li DS, Ainiwaer JL, Sheyhiding I, Zhang Z
and Zhang LW: Identification of key long non-coding RNAs as
competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma.
Eur Rev Med Pharmacol Sci. 20:2285–2295. 2016.PubMed/NCBI
|
|
43
|
Seitz AK, Christensen LL, Christensen E,
Faarkrog K, Ostenfeld MS, Hedegaard J, Nordentoft I, Nielsen MM,
Palmfeldt J, Thomson M, et al: Profiling of long non-coding RNAs
identifies LINC00958 and LINC01296 as candidate oncogenes in
bladder cancer. Sci Rep. 7:3952017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang Y, Guo JX and Shao ZQ: miR-21 targets
and inhibits tumor suppressor gene PTEN to promote prostate cancer
cell proliferation and invasion: An experimental study. Asian Pac J
Trop Med. 10:87–91. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shen SJ, Zhang YH, Gu XX, Jiang SJ and Xu
LJ: Yangfei Kongliu Formula, a compound Chinese herbal medicine,
combined with cisplatin, inhibits growth of lung cancer cells
through transforming growth factor-β1 signaling pathway. J Integr
Med. 15:242–251. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Brattain MG, Markowitz SD and Willson JK:
The type II transforming growth factor-beta receptor as a
tumor-suppressor gene. Curr Opin Oncol. 8:49–53. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chowdhury S, Ammanamanchi S and Howell GM:
Epigenetic targeting of transforming growth factor β receptor II
and implications for cancer therapy. Mo Cell Pharmacol. 1:57–70.
2009. View Article : Google Scholar
|
|
48
|
Bonci D and De Maria R: miR-15/miR-16
loss, miR-21 upregulation, or deregulation of their target genes
predicts poor prognosis in prostate cancer patients. Mol Cell
Oncol. 3:e11097442016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Guan Y, Wu Y, Liu Y, Ni J and Nong S:
Association of microRNA-21 expression with clinicopathological
characteristics and the risk of progression in advanced prostate
cancer patients receiving androgen deprivation therapy. Prostate.
76:986–993. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mishra S, Deng JJ, Gowda PS, Rao MK, Lin
CL, Chen CL, Huang T and Sun LZ: Androgen receptor and microRNA-21
axis downregulates transforming growth factor beta receptor II
(TGFBR2) expression in prostate cancer. Oncogene. 33:4097–4106.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ling H, Fabbri M and Calin GA: MicroRNAs
and other non-coding RNAs as targets for anticancer drug
development. Nat Rev Drug Discov. 12:847–865. 2013. View Article : Google Scholar : PubMed/NCBI
|