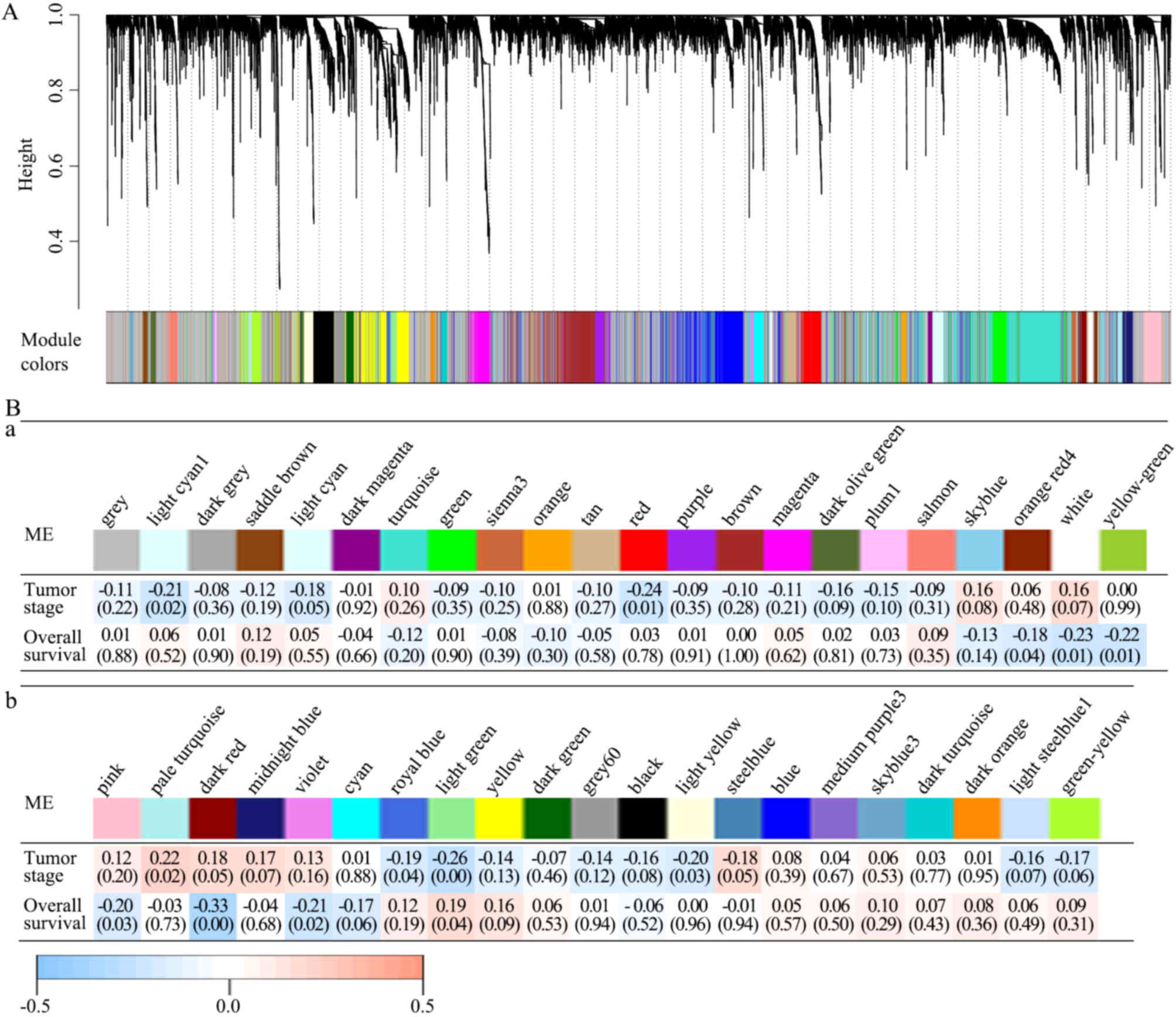
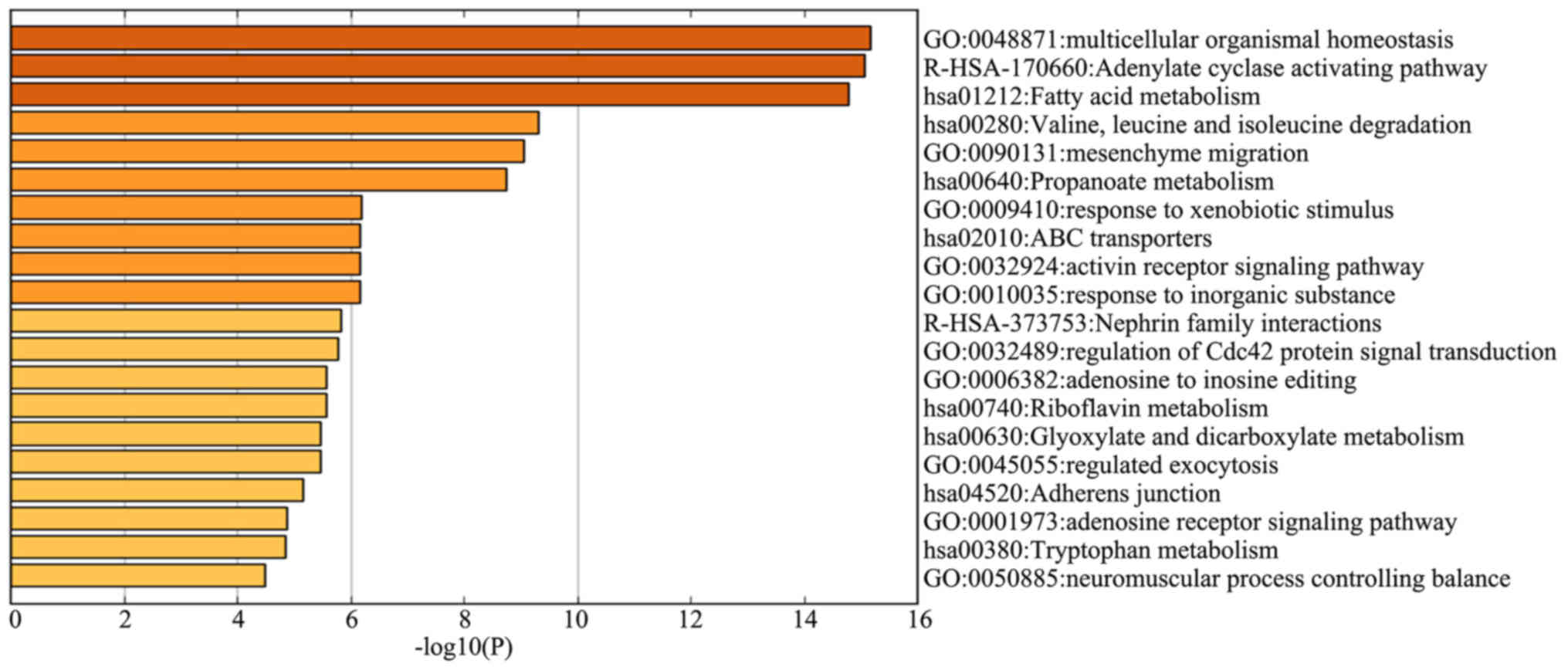
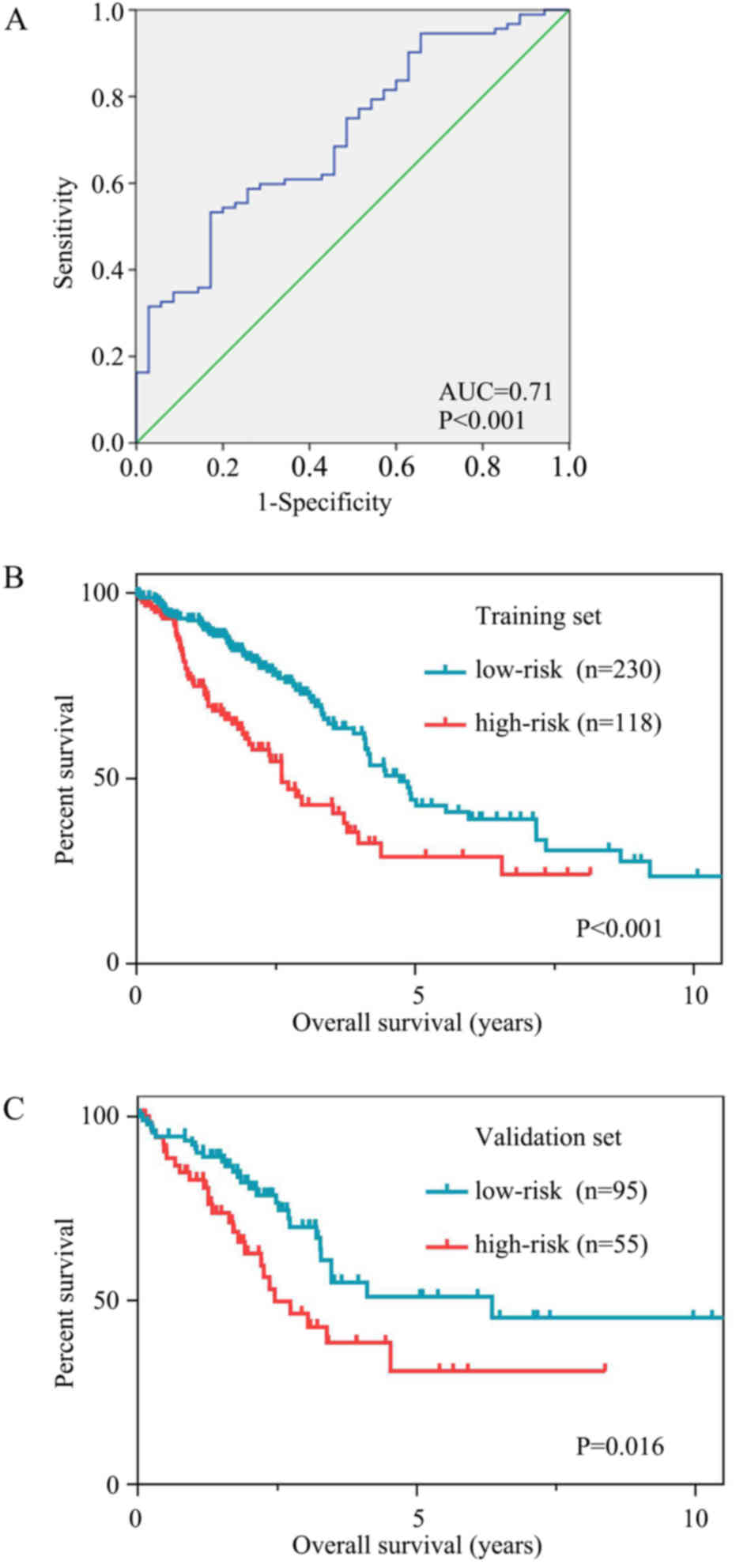
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000–14 (CONCORD-3): Analysis of individual records for 37 513 025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sanchez-Salcedo P, Berto J, de-Torres JP,
Campo A, Alcaide AB, Bastarrika G, Pueyo JC, Villanueva A,
Echeveste JI, Lozano MD, et al: Lung cancer screening: Fourteen
year experience of the Pamplona early detection program (P-IELCAP).
Arch Bronconeumol. 51:169–176. 2015.(In English, Spanish).
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vansteenkiste J, Crinò L, Dooms C,
Douillard JY, Faivre-Finn C, Lim E, Rocco G, Senan S, Van Schil P,
Veronesi G, et al: 2nd ESMO Consensus conference on lung cancer:
Early-stage non-small-cell lung cancer consensus on diagnosis,
treatment and follow-up. Ann Oncol. 25:1462–1474. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Goldstraw P, Chansky K, Crowley J,
Rami-Porta R, Asamura H, Eberhardt WE, Nicholson AG, Groome P,
Mitchell A, Bolejack V, et al: The IASLC lung cancer staging
project: Proposals for revision of the TNM stage groupings in the
forthcoming (Eighth) edition of the TNM classification for lung
cancer. J Thorac Oncol. 11:39–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chansky K, Sculier JP, Crowley JJ, Giroux
D, Van Meerbeeck J, Goldstraw P; International Staging Committee, ;
Participating Institutions: The international association for the
study of lung cancer staging project: Prognostic factors and
pathologic TNM stage in surgically managed non-small cell lung
cancer. J Thorac Oncol. 4:792–801. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cancer Genome Atlas Research Network, ;
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The cancer genome
atlas pan-cancer analysis project. Nat Genet. 45:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Allen JD, Xie Y, Chen M, Girard L and Xiao
G: Comparing statistical methods for constructing large scale gene
networks. PLoS One. 7:e293482012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo L, Zhang K and Bing Z: Application of
a co-expression network for the analysis of aggressive and
non-aggressive breast cancer cell lines to predict the clinical
outcome of patients. Mol Med Rep. 16:7967–7978. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mao Q, Zhang L, Zhang Y, Dong G, Yang Y,
Xia W, Chen B, Ma W, Hu J, Jiang F and Xu L: A network-based
signature to predict the survival of non-smoking lung
adenocarcinoma. Cancer Manag Res. 10:2683–2693. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen J, Wang X, Hu B, He Y, Qian X and
Wang W: Candidate genes in gastric cancer identified by
constructing a weighted gene co-expression network. PeerJ.
6:e46922018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tibshirani R: The lasso method for
variable selection in the Cox model. Stat Med. 16:385–395. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tibshirani R: Regression shrinkage and
selection via the lasso. J R Stat Soc Ser A Stat Soc. 73:273–282.
2011. View Article : Google Scholar
|
|
16
|
Gui J and Li H: Penalized Cox regression
analysis in the high-dimensional and low-sample size settings, with
applications to microarray gene expression data. Bioinformatics.
21:3001–3008. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Langfelder P, Zhang B and Horvath S:
Defining clusters from a hierarchical cluster tree: The dynamic
tree cut package for R. Bioinformatics. 24:719–720. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tripathi S, Pohl MO, Zhou Y,
Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che
J, Mulder LC, et al: Meta- and orthogonal integration of influenza
‘OMICs’ data defines a role for UBR4 in virus budding. Cell Host
Microbe. 18:723–735. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cox DR: Regression models and
life-tablesSpringer; New York: 1992
|
|
21
|
Pasanen L, Holmström L and Sillanpää MJ:
Bayesian LASSO, scale space and decision making in association
genetics. PLoS One. 10:e01200172015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bracht JWP, Mayo-de-Las-Casas C, Berenguer
J, Karachaliou N and Rosell R: The present and future of liquid
biopsies in non-small cell lung cancer: Combining four biosources
for diagnosis, prognosis, prediction, and disease monitoring. Curr
Oncol Rep. 20:702018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao K, Li Z and Tian H: Twenty-gene-based
prognostic model predicts lung adenocarcinoma survival. OncoTargets
Ther. 11:3415–3424. 2018. View Article : Google Scholar
|
|
24
|
Langfelder P and Horvath S: WGCNA package
FAQ. https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/faq.html2017
December 24–2017
|
|
25
|
Friedrich MJ: Going with the flow: The
promise and challenge of liquid biopsies. JAMA. 318:1095–1097.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Siravegna G, Marsoni S, Siena S and
Bardelli A: Integrating liquid biopsies into the management of
cancer. Nat Rev Clin Oncol. 14:531–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fagerberg L, Hallström BM, Oksvold P,
Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S,
Danielsson A, Edlund K, et al: Analysis of the human
tissue-specific expression by genome-wide integration of
transcriptomics and antibody-based proteomics. Mol Cell Proteomics.
13:397–406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiao J, Hong S, Zhang J, Ma L, Sun Y,
Zhang D, Shen B and Zhu C: Opsin3 sensitizes hepatocellular
carcinoma cells to 5-fluorouracil treatment by regulating the
apoptotic pathway. Cancer Lett. 320:96–103. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yoshimoto T, Morine Y, Takasu C, Feng R,
Ikemoto T, Yoshikawa K, Iwahashi S, Saito Y, Kashihara H, Akutagawa
M, et al: Blue light-emitting diodes induce autophagy in colon
cancer cells by Opsin 3. Ann Gastroenterol Surg. 2:154–161. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang B, Yan S, Yan J, Li Y, Khurwolah MR,
Wang L and Chen Z: A study of the association of rs12040273 with
susceptibility and severity of coronary artery disease in a Chinese
Han population. BMC Cardiovasc Disord. 18:102018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Willer CJ, Sanna S, Jackson AU, Scuteri A,
Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS,
Stringham HM, et al: Newly identified loci that influence lipid
concentrations and risk of coronary artery disease. Nat Genet.
40:161–169. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kathiresan S, Melander O, Guiducci C,
Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF,
Havulinna AS, et al: Six new loci associated with blood low-density
lipoprotein cholesterol, high-density lipoprotein cholesterol or
triglycerides in humans. Nat Genet. 40:189–197. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin MC, Huang MJ, Liu CH, Yang TL and
Huang MC: GALNT2 enhances migration and invasion of oral squamous
cell carcinoma by regulating EGFR glycosylation and activity. Oral
Oncol. 50:478–484. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Imielinski M, Berger AH, Hammerman PS,
Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M,
Sivachenko A, et al: Mapping the hallmarks of lung adenocarcinoma
with massively parallel sequencing. Cell. 150:1107–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee SY, Meier R, Furuta S, Lenburg ME,
Kenny PA, Xu R and Bissell MJ: FAM83A confers EGFR-TKI resistance
in breast cancer cells and in mice. J Clin Invest. 122:3211–3220.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Y, Dong X, Yin Y, Su Y, Xu Q, Zhang Y,
Pang X, Zhang Y and Chen W: BJ-TSA-9, a novel human tumor-specific
gene, has potential as a biomarker of lung cancer. Neoplasia.
7:1073–1080. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu L, Liao GQ, He P, Zhu H, Liu PH, Qu
YM, Song XM, Xu QW, Gao Q, Zhang Y, et al: Detection of circulating
cancer cells in lung cancer patients with a panel of marker genes.
Biochem Biophys Res Commun. 372:756–760. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Y, Xiao X, Ji X, Liu B and Amos CI:
RNA-seq analysis of lung adenocarcinomas reveals different gene
expression profiles between smoking and nonsmoking patients. Tumour
Biol. 36:8993–9003. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lauvrak SU, Munthe E, Kresse SH, Stratford
EW, Namløs HM, Meza-Zepeda LA and Myklebost O: Functional
characterisation of osteosarcoma cell lines and identification of
mRNAs and miRNAs associated with aggressive cancer phenotypes. Br J
Cancer. 109:2228–2236. 2013. View Article : Google Scholar : PubMed/NCBI
|