Introduction
The eukaryotic genome transcribes a diverse range of
RNA molecules, from long protein-coding mRNAs to non-coding RNAs
(ncRNAs) (1). Approximately 70% of
the human genome encodes ncRNAs, while protein-coding genes account
for <2% of transcribed RNA (2–5). ncRNAs
can be divided into short ncRNAs (<200 nucleotides in length)
and long ncRNAs (lncRNAs; >200 nucleotides in length) (2–4). It is
generally thought that lncRNAs lack protein-coding capabilities
and, therefore, they were originally regarded as inconsequential
transcriptional noise (2). With
continuous advances in sequencing technology and large-scale genome
sequencing projects, lncRNAs have become a major focus of research.
LncRNAs have been reported to serve extensive roles in various
cellular and physiological processes, such as chromatin dynamics,
and transcriptional and post-transcriptional processes (6–8).
Numerous studies have demonstrated that dysregulation of lncRNAs
may be involved in the pathological process of several human
diseases, including cancer (9,10),
cardiovascular disease (11) and
diseases of the central nervous system (12). Through epigenetic modifications,
chromatin remodeling and microRNA (miRNA) sponging, lncRNAs
modulate cell proliferation, apoptosis, migration and invasion
(13–15). Due to the tissue-specific expression
and high stability of lncRNAs (which allows them to be detected in
body fluids), they may be used as biomarkers for the effective
diagnosis, prognosis and monitoring of disease progression
(16).
Taurine up-regulated 1 (TUG1) is a 7,598-nucleotide
lncRNA sequence localized to chromosome 22q12.2 and was originally
identified in a genomic screen of taurine-treated mouse retinal
cells (17,18). TUG1 has been demonstrated to serve
crucial regulatory roles in various cancer-associated biological
processes and may provide a putative novel treatment paradigm in
cancer therapy. The results of two previous meta-analyses have
indicated that increased expression of TUG1 is an unfavorable
predictor of survival in human cancer, and that TUG1 is closely
associated with increased tumor size, advanced pathological stage
and distant metastasis (19,20). In addition, TUG1 is ubiquitously
expressed and has been implicated in different cancer types. TUG1
has been demonstrated to be upregulated in renal cell carcinoma
(21), ovarian cancer (22), bladder urothelial carcinoma (23), osteosarcoma (24), oral squamous cell carcinoma (25), esophageal squamous cell carcinoma
(26), hepatocellular carcinoma
(HCC) (27), intrahepatic
cholangiocarcinoma (28), glioma
(29), cervical cancer (30), endometrial cancer (31), pancreatic cancer (32), breast cancer (33), bladder cancer (34), colorectal cancer (CRC) (35), small cell lung cancer (SCLC)
(36), melanoma (37), thyroid cancer (38), gallbladder carcinoma (39) and gastric cancer (GC) (40). By contrast, TUG1 expression has been
observed to be downregulated in non-SCLC (NSCLC) (41), triple-negative breast cancer (TNBC)
(42) and glioma (43). In addition, previous studies have
reported that aberrant expression of TUG1 may affect gene
expression through diverse mechanisms, which affect various
biological processes, including cell proliferation, invasion,
apoptosis, differentiation, migration, drug resistance, radiation
resistance, angiogenesis, mitochondrial bioenergetics,
epithelial-mesenchymal transition (EMT) and blood tumor barrier
permeability regulation (44–49). By
conducting an extensive search of the relevant literature, this
review summarizes the key findings of previous studies and
elaborates on the cellular functions and mechanisms of TUG1
regulation, as well as its role in human cancer, particularly
focusing on its role in cell proliferation, invasion, apoptosis,
migration and drug resistance.
Molecular mechanisms of TUG1 in cancer
Accumulating evidence suggests that the molecular
function of lncRNAs is important in epigenetic regulation and that
their molecular mechanisms may be diverse (44,50).
Recent studies have suggested that TUG1 is involved in gene
regulation through a variety of mechanisms; primarily by
functioning as a miRNA sponge and via interacting with polycomb
repressive complex 2 (PRC2) (41,51).
TUG1 functions as a competing endogenous RNA (ceRNA) to target
miRNAs, thereby inhibiting their biological functions (28,29,31,37,42).
This leads to changes in the expression level of downstream target
genes. Khalil et al (51)
confirmed that TUG1 recruits and binds with PRC2. PRC2 possesses
methyltransferase activity and consists of enhancer of zeste
homologue 2 (EZH2), suppressor of zeste 12 protein homolog (SUZ12),
embryonic ectoderm development (EED) and retinoblastoma-associated
protein 46/48. This complex catalyzes the di- and tri-methylation
of lysine residue 27 on histone 3 (52,53). The
binding of TUG1 to PRC2 directs the concomitant genomic DNA to
polycomb bodies leading to epigenetic silencing. The various
molecular mechanisms of TUG1 are summarized in the following
text.
TUG1 in cancer cell proliferation and
apoptosis
It has been demonstrated that TUG1 is capable of
promoting both the proliferation and apoptosis of cells (18,21,24,26,30–34).
Furthermore, TUG1 can exert the same biological function through
the regulation of different target genes in different cell types
(27,28,31,36–39).
Investigating the interactions between lncRNAs and
miRNAs is a popular focus of current ncRNA research (54). As aforementioned, TUG1 exerts its
function in cancer cell proliferation and apoptosis by serving as
an miRNA sponge. In papillary thyroid cancer cells, TUG1 functions
as a ceRNA to sequester miR-145, which in turn leads to an increase
in the expression of the miR-145 target, zinc finger E-box binding
homeobox 1 (ZEB1). Therefore, TUG1 mediates increased cell
proliferation via regulating the miR-145/ZEB1 signaling pathway
(38). In cervical cancer, TUG1
exerts an oncogenic role by binding to miR-138-5p and resulting in
its neutralization. Sirtuin 1 (SIRT1) competes with miR-138-5p and,
thus, is upregulated by TUG1. Highly expressed SIRT1 promotes the
expression of c-myc, β-catenin and cyclin D1, and inhibits the
expression of E-cadherin. This leads to activation of the
Wnt/β-catenin signaling pathway, and subsequent inhibition of
apoptosis and promotion of cervical cancer cell proliferation and
invasion (55).
A previous study reported that TUG1 serves an
oncogenic role in human malignant melanoma via upregulating
astrocyte elevated gene-1 (AEG1) expression by sponging miR-129-5p
(37). AEG1 serves an important role
in tumorigenesis via the phosphatidylinositol-4,5-bisphosphate
3-kinase (PI3K)/AKT (56) and Wnt
signaling pathways (57). Activation
of the TUG1/miR-129-5p/AEG-1 axis in malignant melanoma cells leads
to the inhibition of proliferation and increased apoptosis
(37). Suppression of TUG1 inhibits
the expression of B-cell lymphoma-2 (Bcl-2), matrix
metallopeptidase-9 and cyclin D1, and increases cleaved caspase 3
levels, via modulating the expression of AEG1 in melanoma cells. As
a consequence, overexpression of TUG1 leads to tumorigenesis.
Therefore, suppression of TUG1 expression may inhibit tumorigenesis
and the proliferation of malignant human melanoma cells. Yan et
al (58) reported that miR-219
suppresses the proliferation of oral squamous cell carcinoma cells
by regulating the expression of formin-like 2 (FMNL2). Furthermore,
it was demonstrated that TUG1 sponges miR-219 to regulate its
expression in oral squamous cell carcinoma. In addition, Zhao and
Ren (59) indicated that TUG1
promotes cell proliferation and inhibits apoptosis of breast cancer
cells by functioning as an endogenous sponge of miR-9, thus
affecting the expression of the miR-9 target gene,
methylenetetrahydrofolate dehydrogenase
(NADP+-dependent) 2. In gallbladder carcinoma, TUG1
increases cell proliferation by negatively regulating miR-300
(39); however, the downstream
target gene of miR-300 remains unclear. miRNA-26a binding sites
have been identified in the TUG1 sequence, and a negative
correlation between TUG1 and miR-26a expression has been observed
in prostate cancer (60). Therefore,
the mechanisms underlying TUG1-mediated cell proliferation and
apoptosis may be based, in part, on its negative regulation of
miR-26a. In addition, TUG1 and miR-382 have been demonstrated to
negatively regulate each other. The TUG1/miR-382/EZH2 signaling
pathway has been identified as a ceRNA regulatory network involved
in the proliferation of pancreatic cancer cells (61).
TUG1 has been reported to sequester miR-382 and
downregulate its expression in pancreatic cancer (61). Similarly, TUG1 has been observed to
interact with additional miRNAs, including miR-29c, miR-142 and
miR-145 in bladder cancer (62–64).
EZH2, a downstream target gene of miR-382, miR-142 and miR-145, is
positively regulated by TUG1 (61,63,64).
Overexpression of TUG1 promotes the proliferation and induces the
apoptosis of HCC cells. Furthermore, silencing of TUG1 reduced the
expression of the sonic hedgehog protein, known to be associated
with the Hedgehog (Hh) signaling pathway, and led to an increase in
the expression of miR-132 in hepatocellular carcinoma cells
(65). As reported previously,
overexpression of miR-132 reduces cell proliferation and promotes
apoptosis. The Hh gene was initially identified in
Drosophila in 1980, and Hh has been implicated in the
classical signaling pathway activated during embryonic development
and cell differentiation (66).
Considering these previous observations, it is thought that TUG1
may function as an oncogene and induce tumorigenesis by regulating
the Hh signaling pathway. In addition, TUG upregulates the level of
Janus kinase 2 epigenetically through binding to miR-144, thus
promoting HCC cell proliferation (67).
In addition to the indirect regulation of ceRNAs,
studies published to date have demonstrated that high TUG1
expression in osteosarcoma results in sponging 9 miRNAs, including
miR-212-3p, miR-132-3p, miR-144-3p, miR-153, miR-9-5p and
miR-219a-5p, which affects cell proliferation and apoptosis
(68–73). Xie et al (68) demonstrated that TUG1 upregulated
forkhead box A1 (FOXA1) expression and FOXA1-mediated cell
proliferation and apoptosis by functioning as a ceRNA through
miR-212-3p. In addition, TUG1 was observed to affect the expression
of sex-determining region Y-box 4 by functioning as a ceRNA of
miR-132-3p (69). TUG1 shares
miR-144-3p-response elements with EZH2, which prevents it from
undergoing miR-144-3p-mediated degradation (70). Furthermore it has been shown that
TUG1 contributes to osteosarcoma tumorigenesis by sponging miR-153
(71). Mechanistically, TUG1 binds
to miR-9-5p and thereby inhibits its activity. Concordantly, TUG1
upregulates the miR-9-5p downstream target gene, POU class 2
homeobox 1, which is involved in cell proliferation, colony
formation, cell cycle arrest and apoptosis (72). A recent study has demonstrated that
high TUG1 expression in osteosarcoma leads to the downregulation of
miR-219a-5p expression via an endogenous sponge adsorption
mechanism. This leads to further upregulation of PI3KCA and
activation of the AKT signaling pathway, which promotes cell
proliferation (73).
A major role of TUG1 is regulating gene expression
and influencing cell proliferation via binding to PRC2. Modulation
of TUG1 alters the level of multiple genes in various cancer cells;
the majority of which are PRC2-dependent. A diverse number of genes
are targeted by TUG1-induced PRC2, including CUGBP Elav-like family
member 1 (CELF1), homeobox B7 (HOXB7), LIM domain kinase 2b
(LIMK2b), p57, Kruppel-like factor 2 (KLF2) and Bax; a number of
these are positive regulators of cell proliferation and apoptosis
(27,36,41,74–78). Lin
et al (41) demonstrated that
the expression levels of TUG1 are prominently downregulated in
NSCLC tumors. In addition, knockdown of TUG1 significantly
increases the proliferation of NSCLC cells. TUG1 affects NSCLC cell
proliferation by negatively regulating CELF1. Of particular note is
that chromatin immunoprecipitation assays have confirmed that
EZH2/EED bind to the promoter region of CELF1 at 992 bp upstream of
the transcription start site. Consequently, TUG1 binds with PRC2
and occupies the promoter region of CELF1. An additional study
reported that TUG1 is required to activate PRC2 by occupying its
binding site on HOXB7 (a known oncogene), thereby epigenetically
modulating its expression and promoting NSCLC cell proliferation
(74,75). In SCLC, TUG1 downregulates LIMK2b
expression by binding with EZH2 to promote SCLC cell proliferation
(36). In addition, TUG1 recruits
EZH2 to target gene promoters and epigenetically represses the
expression of cyclin-dependent protein kinase inhibitors, including
p15, p16, p21, p27 and p57 in GC. This leads to TUG1-mediated
alterations in the proliferation of GC cells and cell cycle
progression (76). Ultimately, the
results of these studies indicate that TUG1 may function as a
regulator of cell proliferation. In addition, knockdown of TUG1 in
a HCC cell line, Hep3B, led to inhibition of cell proliferation and
induction of cell apoptosis in vitro (27). TUG1 recruits PRC2 to the KLF2 gene
locus, which leads to the direct binding of EZH2 to its promoter
region and inactivation of KLF2 expression. The low expression
levels of KLF2 (a known tumor suppressor gene) results in reduced
cell apoptosis and permits increased proliferation of HCC cells
(27,77). TUG1 regulates cancer cell apoptosis
in lung adenocarcinoma via inhibition of Bax, which is a
pro-apoptotic protein that interacts with EZH2 (78).
In ovarian epithelial cancer cells, TUG1 has been
reported to inhibit apoptosis and promote cell proliferation by
upregulating the expression of aurora kinase A, which has been
shown to be implicated in the carcinogenic processes of multiple
human cancer types (79–81). In addition, knockdown of TUG1 in oral
squamous cell carcinoma cells leads to decreased proliferation
in vitro (25). Reverse
transcription-quantitative PCR and western blotting results
indicated that knockdown of TUG1 leads to downregulation of
Wnt/β-catenin signaling-associated genes, such as β-catenin, cyclin
D1 and c-myc (25). In general, TUG1
promotes cell growth and proliferation via targeting the
Wnt/β-catenin signaling pathway.
Recent studies have proposed that TUG1 exerts
anti-apoptotic effects in CRC (82),
bladder cancer (23), ovarian cancer
(22), oral squamous cell carcinoma
(25) and cervical cancer (30) cells by regulating the expression of
apoptosis-associated mediators, including caspase 3, caspase 9 and
Bcl-2. Yin et al (83)
demonstrated that the level of TUG1 expression in pancreatic
tissues is significantly higher than that in other organ
tissues.
TUG1 in cancer migration and invasion
It has been reported that EMT serves a significant
role in mediating migration and invasion (84). During EMT, the expression of
epithelial cell markers, including E-cadherin, cytokeratins and
claudins, is downregulated. By contrast, the expression of
vimentin, N-cadherin, fibronectin and mucin 1, which function as
mesenchymal markers, is upregulated (85). Recently, an association between TUG1
and EMT was identified. When TUG1 expression is knocked down in
ovarian cancer cells, E-cadherin expression is high, while vimentin
and N-cadherin levels are downregulated. Simultaneously, the
proliferation and metastasis of ovarian cancer cell lines are
markedly inhibited. These results suggest that TUG1 may participate
in modulating cell migration and invasion via EMT (22). Similarly, in bladder and cervical
cancer, TUG1 was demonstrated to promote cell invasion by inducing
EMT (30,34). In addition, overexpression of TUG1
promoted the migration, invasion and EMT of thyroid cancer cell
lines, and these effects were primarily dependent on the
TUG1-mediated regulation of the miR-145/ZEB1 signaling pathway
(38). Furthermore, it was
demonstrated that knockdown of TUG1 markedly inhibited the invasion
and migration capabilities of prostate cancer cells (61). Mechanistically, silencing of TUG1
promotes changes in cell morphology from a mesenchymal to an
epithelial phenotype. TUG1 facilitates pancreatic cancer cell
invasion by stimulating ZEB2-mediated EMT (61). In addition, TUG1 has been reported to
accelerate EMT by promoting the expression of KIAA1199 (also known
as cell migration-inducing hyaluronidase 1) by reducing miR-600
expression (86). It was
demonstrated that high levels of miR-600 inhibited CRC migration,
invasion and the expression of EMT-associated proteins in
vitro. However, KIAA1199 was indirectly upregulated by TUG1
(86).
Following confirmation that miR-219a-5p targets TUG1
using a luciferase assay, it was demonstrated that TUG1 regulates
the expression of PIK3CA and activates the AKT signaling pathway to
promote the migration and invasion of osteosarcoma cells (87). In addition, TUG1 was observed to
downregulate the expression of miR-335-5p in osteosarcoma via an
endogenous sponge adsorption mechanism, which led to further
upregulation of ρ-associated coiled-coil containing protein kinase
1 and increased cell migration and invasion (73). A recent study reported that TUG1
induces the invasion of oral squamous cell carcinoma cells via
repression of miR-219 or regulation of FMNL2 (58). In addition, Zhao et al
(61) demonstrated that TUG1
promotes the migration and EMT of pancreatic cancer cells via
downregulation of miR-382 and regulation of its target, EZH2
(61). Furthermore, miR-144 has been
demonstrated to bind directly with TUG1. Recent studies have
investigated whether TUG1 may function as a ceRNA and bind to miRNA
response elements in miR-144, thus inhibiting its expression in GC
and HCC (67,88). Together, these studies provide
evidence indicating a significant correlation between increased
TUG1 expression and invasion. Ji et al (88) demonstrated that TUG1 is markedly
upregulated in GC tissues and that silencing of TUG1 leads to the
inhibition of c-Met and the inhibition of invasion capabilities of
SGC-7901 cells. In HCC, TUG1 competes with miR-144 for binding to
the 3′-untranslated region of Janus kinase 2 (JAK2), thereby
activating the JAK2/signal transducer and activator of
transcription 3 signaling pathway and increasing cell migration
in vitro and in vivo (67). He et al (89) demonstrated that high TUG1 levels were
positively associated with cell invasion in HCC. TUG1 directly
interacts with miR-142-3p. In addition, miR-142-3p suppresses HCC
cell invasion and metastasis by targeting ZEB1. Silencing of TUG1
consequently reduces the expression levels of ZEB1 and
EMT-associated proteins. Moreover, TUG1 acts as an oncogene in HCC
progression by binding with miR-142-3p and abrogating its
tumor-suppressive function. Furthermore, it has been reported that
the TUG1/PRC2/p21/miR-455-3p/AMPKβ2 axis regulates hexokinase 2
(HK2) to mediate HCC cell invasion (90). TUG1 downregulates p21 expression via
upregulation of EED expression, which leads to reduced p21-E2F1
interaction and enhanced miR-455-3p expression. The expression of
AMPKβ2 is inhibited by upregulation of miR-455-3p and thereby HK2
expression is increased.
TUG1 in cancer drug resistance
Drug resistance is one of the most important reasons
for therapeutic failure in patients with cancer and is a persistent
issue that requires continued investigation. Tang et al
(42) demonstrated that the
expression of miR-197 is elevated in human TNBC tumor tissues and
cell lines (MDA-MB-231 and BT549), while the levels of TUG1 and
nemo-like kinase (NLK) are decreased. miR-197 confers cisplatin
resistance in TNBC by inhibiting NLK. However, TUG1 functions as an
endogenous sponge of miR-9-5p. Increased expression of TUG1
sensitizes TNBC cells to very low concentrations of cisplatin.
Therefore, the TUG1/miR-197/NLK signaling pathway is likely to be a
novel and promising therapeutic target for patients with TNBC.
TUG1 has been implicated in the resistance of human
CRC cells to methotrexate (MTX) via the miR-186/cytoplasmic
polyadenylation element binding protein 2 (CPEB2) axis (91). MTT assays have demonstrated that
miR-186 re-sensitizes CRC cells to MTX. By contrast, CPEB2 enhances
the resistance of CRC cells to MTX. TUG1 functions as a ceRNA to
upregulate CPEB2 by sponging miR-186. Overexpression of TUG1
significantly enhances MTX resistance in CRC cells, which suggests
that TUG1 may present a putative therapeutic target in CRC.
Regulation and dysregulation of TUG1
The expression level of TUG1 and the association
between expression and survival probability in several common
tumors were searched using The Cancer Genome Atlas database,
including HCC, melanoma, osteosarcoma, renal cell carcinoma,
bladder cancer, CRC, GC and triple negative breast cancer. The
figures of the statistical results were obtained from this database
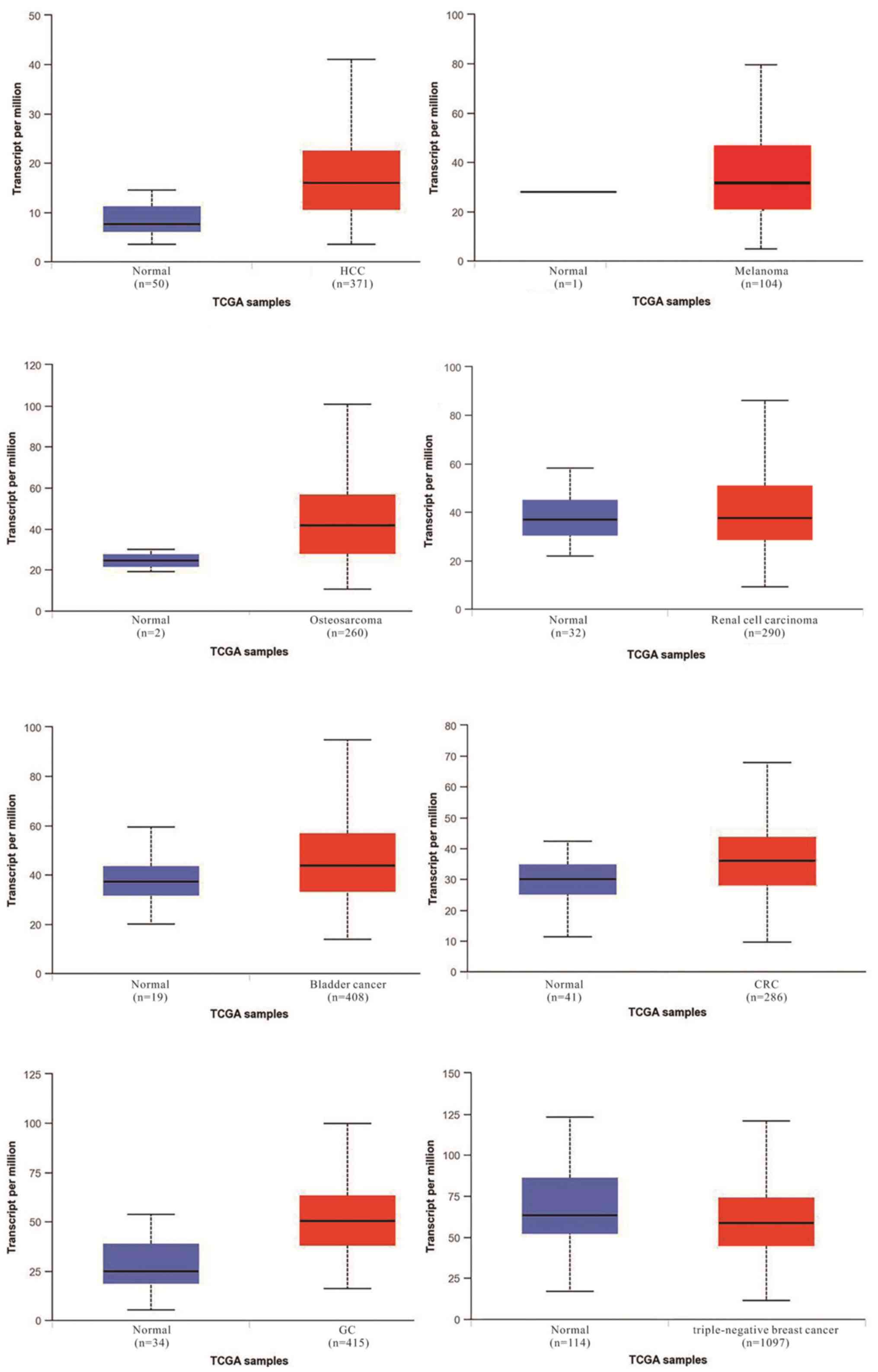
(Figs. 1 and 2). According to The Cancer Genome Atlas
database, there is a marked difference in the expression of TUG1
between normal and tumor tissues (Fig.
1). TUG1 serves both oncogenic and tumor suppressive functions
depending on the type of cancer. As an oncogene, aberrant
upregulation of this lncRNA in different cancer types compared with
their noncancerous counterparts has been observed in HCC, melanoma,
osteosarcoma, renal cell carcinoma, bladder cancer, CRC and GC. By
contrast, other reports have observed downregulation of TUG1
in TNBC. However, it has been demonstrated that high TUG1
expression may indicate a poor prognosis in the majority of cancer
types (Fig. 2).
A recent bioinformatics study demonstrated that the
TUG1 promoter region harbors five SP1 binding sites, and it has
been reported that SP1 directly binds to the TUG1 promoter region
to upregulate TUG1 expression (27).
In addition, Zhang et al (74) demonstrated that TUG1 is a direct
transcriptional target of p53. p53 binds to its putative response
element in the promoter region of TUG1. The p53 tumor suppressor
gene is associated with the occurrence and development of various
cancer types, such as breast cancer (92), bladder cancer (93), liver cancer (94) and glioma (95). It is possible that TUG1 may be
closely associated with p53 and cancer; however, further research
to verify this hypothesis is required. A previous study involving
RNA-sequence and lncRNA microarray analyses has demonstrated that
TUG1 is commonly downregulated in glioma following inhibition of
Notch signaling. Notch1 was identified to bind with recombination
signal binding protein for immunoglobulin κ J region motifs located
upstream of the transcription start site of TUG1. TUG1 expression
is induced by Notch signaling (46).
TUG1 has been reported to be induced by forkhead box
M1 (FOXM1) in osteosarcoma cells (73). Genome-wide gene expression profiling
of human solid tumors has demonstrated that FOXM1 is one of the
most commonly upregulated genes across different cancer types
(96,97). AKT activates FOXM1 by inducing the
phosphorylation of FOXO3 for protein degradation in osteosarcoma
cells. TUG1 and FOXM1 are concurrently highly expressed in human
osteosarcoma. Consistent with these observations, increased FOXM1
expression was demonstrated to activate the TUG1 promoter in a
dose-dependent manner. In addition, induction of TUG1 by FOXM1 is
diminished when FOXM1 expression levels are silenced by small
interfering RNA (siRNA). These observations suggest that increased
FOXM1 expression by AKT may upregulate the expression level of TUG1
via direct interaction with the TUG1 promoter in osteosarcoma
cells.
Discussion and perspectives
lncRNAs, which are non-protein-coding RNA sequences,
serve important roles in multiple cellular processes. TUG1, as a
novel lncRNA, has been demonstrated to be abnormally expressed in
various human cancer types, and its dysregulation is closely
associated with carcinogenesis and disease progression. Emerging
evidence suggests that TUG1 may be a vital mediator of cell
proliferation, apoptosis, migration, invasion and drug resistance.
Further study of TUG1, which is considered to function as a gene
regulator, may aid in addressing disease etiology. From a molecular
perspective, cancer is a genetic disease that develops due to the
aberrant expression and function of tumor suppressor and oncogenic
genes. TUG1 functions as an oncogene or tumor suppressor depending
on the type of cancer. As such, silencing or upregulating TUG1 may
inhibit the proliferation, migration and invasion of cancer cells
and increase cell apoptosis. The present review summarizes the
mechanisms of TUG1 in mediating these biological processes and
discusses the regulation of TUG1 (Fig.
3).
 | Figure 3.Schematic illustration of TUG1
regulatory mechanisms. TRAF5, TNF receptor-associated factor 5;
FOXM1, forkhead box M1; PRC2, polycomb repressive complex 2; CELF1,
CUGBP Elav-like family member 1; HOXB7, homeobox B7; LIMK2b, LIM
domain kinase 2b; KLF2, Kruppel-like factor 2; AURKA, aurora kinase
A; HK, hexokinase 2; AEG1, astrocyte elevated gene-1; FOXA1,
forkhead box A1; EZH2, enhancer of zeste homologue 2; JAK2, Janus
kinase 2; SIRT1, Sirtuin 1; ZEB1, zinc finger E-box binding
homeobox 1; CPEB2, cytoplasmic polyadenylation element binding
protein 2; KIAA1199, migration inducing hyaluronidase 1; EMT,
epithelial-mesenchymal transition. |
Notwithstanding progress in the field of lncRNA
research in recent years, studies investigating the role of TUG1 in
cancer are still at a very early stage. For instance, Li et
al (43,98) has demonstrated that TUG1 is
significantly reduced in U251 and SHG-44 glioma cells. This has
provided a novel insight for the potential treatment of patients
with glioma by overexpressing TUG1. By contrast, other studies have
reported that TUG1 is upregulated in glioma tissues and cell lines
(29,45) and therefore functions as a putative
oncogene. Therefore, the role of TUG1 in glioma is contradictory
and requires further investigation.
In conclusion, the regulatory network of TUG1 in the
majority of biological processes varies considerably. However, TUG1
appears to mediate these processes primarily by binding with PCR2
to silence downstream target genes and by sponging miRNAs to
promote the expression of target genes. In theory, TUG1 activity
may be inhibited using numerous strategies. One approach may be to
inhibit molecular interactions using small molecule inhibitors that
block specific binding sites. Alternatively, TUG1 may be silenced
using specific siRNAs. Investigation of TUG1 signaling pathways in
cancer cells may uncover numerous novel therapeutic approaches for
cancer treatment in the future. Therefore, it is essential for
future research to focus on understanding the underlying molecular
mechanisms of TUG1.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by the
grants from the National Natural Science Foundation of China (grant
no. 81360032) and the Jiangxi Natural Science Foundation (grant no.
20161BAB205206).
Availability of data and materials
The datasets generated during the current study are
available in the Cancer Genome Atlas repository, (https://portal.gdc.cancer.gov).
Authors' contributions
HZ and LS conceived and designed the review. HZ
drafted the manuscript. LS participated in the manuscript's
revision and provided suggestions for important content. FW
proposed concepts, revised the article and obtained funding. All
the authors approved the final version of manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Reik W: Evolution and functions of long
noncoding RNAs. Cell. 136:629–641. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wahlestedt C: Targeting long non-coding
RNA to therapeutically upregulate gene expression. Nat Rev Drug
Discov. 12:433–446. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ling H, Fabbri M and Calin GA: MicroRNAs
and other non-coding RNAs as targets for anticancer drug
development. Nat Rev Drug Discov. 12:847–865. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Du Z, Sun T, Hacisuleyman E, Fei T, Wang
X, Brown M, Rinn JL, Lee MG, Chen Y, Kantoff PW and Liu XS:
Integrative analyses reveal a long noncoding RNA-mediated sponge
regulatory network in prostate cancer. Nat Commun. 7:109822016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bhan A and Mandal SS: LongNoncoding RNAs:
Emerging stars in gene regulation, epigenetics and human disease.
ChemMedChem. 9:1932–1956. 2015. View Article : Google Scholar
|
|
7
|
Heo JB, Lee YS and Sung S: Epigenetic
regulation by long noncoding RNAs in plants. Chromosome Res.
21:685–693. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schonrock N, Harvey RP and Mattick JS:
Long noncoding RNAs in cardiac development and pathophysiology.
Circ Res. 111:1349–1362. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Carrieri C, Forrest A, Santoro C,
Persichetti F, Carninci P, Zucchelli S and Gustincich S: Expression
analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1)
during dopaminergic cells' differentiation in vitro and in
neurochemical models of Parkinson's disease. Front Cell Neurosci.
9:1142015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rinn JL: lncRNAs: Linking RNA to
chromatin. Cold Spring Harb Perspect Biol. 6(pii): a0186142014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wilusz JE: Long noncoding RNAs: Re-writing
dogmas of RNA processing and stability. Biochim Biophys Acta.
1859:128–138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kunej T, Obsteter J, Pogacar Z, Horvat S
and Calin GA: The decalog of long non-coding RNA involvement in
cancer diagnosis and monitoring. Crit Rev Clin Lab Sci. 51:344–357.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Young TL, Matsuda T and Cepko CL: The
noncoding RNA Taurine Upregulated gene 1 is required for
differentiation of the murine retina. Curr Biol. 15:501–512. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Q, Geng PL, Yin P, Wang XL, Jia JP
and Yao J: Down-regulation of long non-coding RNA TUG1 inhibits
osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J
Cancer Prev. 14:2311–2315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou Y, Lu Y, Li R, Yan N, Li X and Dai T:
Prognostic role of long non-coding RNA TUG1 expression in various
cancers: A meta-analysis. Oncotarget. 8:100499–100507.
2017.PubMed/NCBI
|
|
20
|
Li N, Shi K, Kang X and Li W: Prognostic
value of long non-coding RNA TUG1 in various tumors. Oncotarget.
8:65659–65667. 2017.PubMed/NCBI
|
|
21
|
Zhang M, Lu W, Huang Y, Shi J, Wu X, Zhang
X, Jiang R, Cai Z and Wu S: Downregulation of the long noncoding
RNA TUG1 inhibits the proliferation, migration, invasion and
promotes apoptosis of renal cell carcinoma. J Mol Histol.
47:421–428. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kuang D, Zhang X, Hua S, Dong W and Li Z:
Long non-coding RNA TUG1 regulates ovarian cancer proliferation and
metastasis via affecting epithelial-mesenchymal transition. Exp Mol
Pathol. 101:267–273. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Han Y, Liu Y, Gui Y, Gui Y and Cai Z: Long
intergenic non-coding RNA TUG1 is overexpressed in urothelial
carcinoma of the bladder. J Surg Oncol. 107:555–559. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yun-Bo F, Xiao-Po L, Xiao-Li L, Guo-Long
C, Pei Z and Fa-Ming T: LncRNA TUG1 is upregulated and promotes
cell proliferation in osteosarcoma. Open Med (Wars). 11:163–167.
2016.PubMed/NCBI
|
|
25
|
Liang S, Zhang S, Wang P, Yang C, Shang C,
Yang J and Wang J: LncRNA, TUG1 regulates the oral squamous cell
carcinoma progression possibly via interacting with Wnt/β-catenin
signaling. Gene. 608:49–57. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu Y, Wang J, Qiu M and Xu L, Li M, Jiang
F, Yin R and Xu L: Upregulation of the long noncoding RNA TUG1
promotes proliferation and migration of esophageal squamous cell
carcinoma. Tumour Biol. 36:1643–1651. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang MD, Chen WM, Qi FZ, Sun M, Xu TP, Ma
P and Shu YQ: Long non-coding RNA TUG1 is up-regulated in
hepatocellular carcinoma and promotes cell growth and apoptosis by
epigenetically silencing of KLF2. Mol Cancer. 14:1652015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zeng B, Ye H, Chen J, Cheng D, Cai C, Chen
G, Chen X, Xin H, Tang C and Zeng J: LncRNA TUG1 sponges miR-145 to
promote cancer progression and regulate glutamine metabolism via
Sirt3/GDH axis. Oncotarget. 8:113650–113661. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cai H, Xue Y, Wang P, Wang Z, Li Z, Hu Y,
Li Z, Shang X and Liu Y: The long noncoding RNA TUG1 regulates
blood-tumor barrier permeability by targeting miR-144. Oncotarget.
6:19759–19779. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu Y, Sun X, Mao C, Guo G, Ye S, Xu J, Zou
R, Chen J, Wang L, Duan P and Xue X: Upregulation of long noncoding
RNA TUG1 promotes cervical cancer cell proliferation and migration.
Cancer Med. 6:471–482. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu L, Chen X, Zhang Y, Hu Y, Shen X and
Zhu W: Long non-coding RNA TUG1 promotes endometrial cancer
development via inhibiting miR-299 and miR-34a-5p. Oncotarget.
8:31386–31394. 2017.PubMed/NCBI
|
|
32
|
Qin CF and Zhao FL: Long non-coding RNA
TUG1 can promote proliferation and migration of pancreatic cancer
via EMT pathway. Eur Rev Med Pharmacol Sci. 21:2377–2384.
2017.PubMed/NCBI
|
|
33
|
Li T, Liu Y, Xiao H and Xu G: Long
non-coding RNA TUG1 promotes cell proliferation and metastasis in
human breast cancer. Breast Cancer. 24:535–543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Iliev R, Kleinova R, Juracek J, Dolezel J,
Ozanova Z, Fedorko M, Pacik D, Svoboda M, Stanik M and Slaby O:
Overexpression of long non-coding RNA TUG1 predicts poor prognosis
and promotes cancer cell proliferation and migration in high-grade
muscle-invasive bladder cancer. Tumour Biol. 37:13385–13390. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sun J, Ding C, Yang Z, Liu T, Zhang X,
Zhao C and Wang J: The long non-coding RNA TUG1 indicates a poor
prognosis for colorectal cancer and promotes metastasis by
affecting epithelial-mesenchymal transition. J Transl Med.
14:422016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Niu Y, Ma F, Huang W, Fang S, Li M, Wei T
and Guo L: Long non-coding RNA TUG1 is involved in cell growth and
chemoresistance of small cell lung cancer by regulating LIMK2b via
EZH2. Mol Cancer. 16:52017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Long J, Menggen Q, Wuren Q, Shi Q and Pi
X: Long Noncoding RNA Taurine-Upregulated Gene1 (TUG1) promotes
tumor growth and metastasis through
TUG1/Mir-129-5p/Astrocyte-Elevated Gene-1 (AEG-1) axis in malignant
melanoma. Med Sci Monit. 24:1547–1559. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lei H, Gao Y and Xu X: LncRNA TUG1
influences papillary thyroid cancer cell proliferation, migration,
and EMT formation through targeting miR-145. Acta Biochim Biophys
Sin. 49:588–597. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ma F, Wang SH, Cai Q, Jin LY, Zhou D, Ding
J and Quan ZW: Long non-coding RNA TUG1 promotes cell proliferation
and metastasis by negatively regulating miR-300 in gallbladder
carcinoma. Biomed Pharmacother. 88:863–869. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Baratieh Z, Khalaj Z, Honardoost MA,
Emadi-Baygi M, Khanahmad H, Salehi M and Nikpour P: Aberrant
expression of PlncRNA-1 and TUG1: Potential biomarkers for gastric
cancer diagnosis and clinically monitoring cancer progression.
Biomark Med. 11:1077–1090. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lin PC, Huang HD, Chang CC, Chang YS, Yen
JC, Lee CC, Chang WH, Liu TC and Chang JG: Long noncoding RNA TUG1,
is downregulated in non-small cell lung cancer and can regulate
CELF1, on binding to PRC2. BMC Cancer. 16:5832016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tang T, Cheng Y, She Q, Jiang Y, Chen Y,
Yang W and Li Y: Long non-coding RNA TUG1 sponges miR-197 to
enhance cisplatin sensitivity in triple negative breast cancer.
Biomed Pharmacother. 107:338–346. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li J, Zhang M, An G and Ma Q: LncRNA TUG1
acts as a tumor suppressor in human glioma by promoting cell
apoptosis. Exp Biol Med (Maywood). 241:644–649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Niland CN, Merry CR and Khalil AM:
Emerging roles for long non-coding RNAs in cancer and neurological
disorders. Front Genet. 3:252012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cai H, Liu X, Zheng J, Xue Y, Ma J, Li Z,
Xi Z, Li Z, Bao M and Liu Y: Long non-coding RNA taurine
upregulated 1 enhances tumor-induced angiogenesis through
inhibiting microRNA-299 in human glioblastoma. Oncogene.
36:318–331. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Katsushima K, Natsume A, Ohka F, Shinjo K,
Hatanaka A, Ichimura N, Sato S, Takahashi S, Kimura H, Totoki Y, et
al: Targeting the Notch-regulated non-coding RNA TUG1 for glioma
treatment. Nat Commun. 7:136162016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Long J, Badal SS, Ye Z, Wang Y, Ayanga BA,
Galvan DL, Green NH, Chang BH, Overbeek PA and Danesh FR: Long
noncoding RNA Tug1 regulates mitochondrial bioenergetics in
diabetic nephropathy. J Clin Invest. 126:4205–4218. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kondo Y, Shinjo K and Katsushima K: Long
non-coding RNAs as an epigenetic regulator in human cancers. Cancer
Sci. 108:1927–1933. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chiu HS, Somvanshi S, Patel E, Chen TW,
Singh VP, Zorman B, Patil SL, Pan Y, Chatterjee SS; Cancer Genome
Atlas Research Network, ; et al: Pan-cancer analysis of lncRNA
regulation supports their targeting of cancer genes in each tumor
context. Cell Rep. 23:297–312.e12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Morales DR, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wahlestedt C: Targeting long non-coding
RNA to therapeutically upregulate gene expression. Nat Rev Drug
Discov. 12:433–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ciferri C, Lander GC, Maiolica A, Herzog
F, Aebersold R and Nogales E: Molecular architecture of human
polycomb repressive complex 2. Elife. 1:e000052012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yoon JH, Abdelmohsen K and Gorospe M:
Functional interactions among microRNAs and long noncoding RNAs.
Semin Cell Dev Biol. 34:9–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhu J, Shi H, Liu H, Wang X and Li F: Long
non-coding RNA TUG1 promotes cervical cancer progression by
regulating the miR-138-5p-SIRT1 axis. Oncotarget. 8:65253–65264.
2017.PubMed/NCBI
|
|
56
|
Xie Y and Zhong DW: AEG-1 is associated
with hypoxia-induced hepatocellular carcinoma chemoresistance via
regulating PI3K/AKT/HIF-1alpha/MDR-1 pathway. EXCLI J. 15:745–757.
2016.PubMed/NCBI
|
|
57
|
He W, He S, Wang Z, Shen H, Fang W, Zhang
Y, Qian W, Lin M, Yuan J, Wang J, et al: Astrocyte elevated
gene-1(AEG-1) induces epithelial-mesenchymal transition in lung
cancer through activating Wnt/β-catenin signaling. BMC Cancer.
15:1072015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yan G, Wang X, Yang M, Lu L and Zhou Q:
Long non-coding RNA TUG1 promotes progression of oral squamous cell
carcinoma through upregulating FMNL2 by sponging miR-219. Am J
Cancer Res. 7:1899–1912. 2017.PubMed/NCBI
|
|
59
|
Zhao XB and Ren GS: LncRNA
taurine-upregulated gene 1 promotes cell proliferation by
inhibiting MicroRNA-9 in MCF-7 cells. J Breast Cancer. 19:349–357.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yang B, Tang X, Wang Z, Sun D, Wei X and
Ding Y: TUG1 promotes prostate cancer progression by acting as a
ceRNA of miR-26a. Biosci Rep. 38(pii): BSR20180677. 2018.
|
|
61
|
Zhao L, Sun H, Kong H, Chen Z, Chen B and
Zhou M: The Lncrna-TUG1/EZH2 axis promotes pancreatic cancer cell
proliferation, migration and EMT phenotype formation through
sponging Mir-382. Cell Physiol Biochem. 42:2145–2158. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Guo P, Zhang G, Meng J, He Q, Li Z and
Guan Y: Upregulation of long non-coding RNA TUG1 promotes bladder
cancer cell proliferation, migration, and invasion by inhibiting
miR-29c. Oncol Res. 26:1083–1091. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu Q, Liu H, Cheng H, Li Y, Li X and Zhu
C: Downregulation of long noncoding RNA TUG1 inhibits proliferation
and induces apoptosis through the TUG1/miR-142/ZEB2 axis in bladder
cancer cells. Onco Targets Ther. 10:2461–2471. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Tan J, Qiu K, Li M and Liang Y:
Double-negative feedback loop between long non-coding RNA TUG1 and
miR-145 promotes epithelial to mesenchymal transition and
radioresistance in human bladder cancer cells. FEBS Lett.
589:3175–3181. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li J, Zhang Q, Fan X, Mo W, Dai W, Feng J,
Wu L, Liu T, Li S, Xu S, et al: The long noncoding RNA TUG1 acts as
a competing endogenous RNA to regulate the Hedgehog pathway by
targeting miR-132 in hepatocellular carcinoma. Oncotarget.
8:65932–65945. 2017.PubMed/NCBI
|
|
66
|
Bakshi A, Chaudhary SC, Rana M, Elmets CA
and Athar M: Basal cell carcinoma pathogenesis and therapy
involving hedgehog signaling and beyond. Mol Carcinog.
56:2543–2557. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lv J, Kong Y, Gao Z, Liu Y, Zhu P and Yu
Z: LncRNA TUG1 interacting with miR-144 contributes to
proliferation, migration and tumorigenesis through activating the
JAK2/STAT3 pathway in hepatocellular carcinoma. Int J Biochem Cell
Biol. 101:19–28. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Xie C, Chen B, Wu B, Guo J and Cao Y:
LncRNA TUG1 promotes cell proliferation and suppresses apoptosis in
osteosarcoma by regulating miR-212-3p/FOXA1 axis. Biomed
Pharmacother. 97:1645–1653. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Li G, Liu K and Du X: Long non-coding RNA
TUG1 promotes proliferation and inhibits apoptosis of osteosarcoma
cells by sponging miR-132-3p and Upregulating SOX4 expression.
Yonsei Med J. 59:226–235. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Cao J, Han X, Qi X, Jin X and Li X: TUG1
promotes osteosarcoma tumorigenesis by upregulating EZH2 expression
via miR-144-3p. Int J Oncol. 51:1115–1123. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang H, Yu Y, Fan S and Luo L: Knockdown
of long noncoding RNA TUG1 inhibits the proliferation and cellular
invasion of osteosarcoma cells by sponging MiR-153. Oncol Res.
26:665–673. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Xie CH, Cao YM, Huang Y, Shi QW, Guo JH,
Fan ZW, Li JG, Chen BW and Wu BY: Long non-coding RNA TUG1
contributes to tumorigenesis of human osteosarcoma by sponging
miR-9-5p and regulating POU2F1 expression. Tumour Biol.
37:15031–15041. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Li Y, Zhang T, Zhang Y, Zhao X and Wang W:
Targeting the FOXM1-regulated long non-coding RNA TUG1 in
osteosarcoma. Cancer Sci. 109:3093–3104. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang EB, Yin DD, Sun M, Kong R, Liu XH,
You LH, Han L, Xia R, Wang KM, Yang JS, et al: P53-regulated long
non-coding RNA TUG1 affects cell proliferation in human non-small
cell lung cancer, partly through epigenetically regulating HOXB7
expression. Cell Death Dis. 5:e12432014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Liao WT, Jiang D, Yuan J, Cui YM, Shi XW,
Chen CM, Bian XW, Deng YJ and Ding YQ: HOXB7 as a prognostic factor
and mediator of colorectal cancer progression. Clin Cancer Res.
17:3569–3578. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhang E, He X, Yin D, Han L, Qiu M, Xu T,
Xia R, Xu L, Yin R and De W: Increased expression of long noncoding
RNA TUG1 predicts a poor prognosis of gastric cancer and regulates
cell proliferation by epigenetically silencing of p57. Cell Death
Dis. 7:e21092016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Shen P, Sun J, Xu G, Zhang L, Yang Z, Xia
S, Wang Y, Liu Y and Shi G: KLF9, a transcription factor induced in
flutamide-caused cell apoptosis, inhibits AKT activation and
suppresses tumor growth of prostate cancer cells. Prostate.
74:946–958. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Liu H, Zhou G, Fu X, Cui H, Pu G, Xiao Y,
Sun W, Dong X, Zhang L, Cao S, et al: Long noncoding RNA TUG1 is a
diagnostic factor in lung adenocarcinoma and suppresses apoptosis
via epigenetic silencing of BAX. Oncotarget. 8:101899–101910.
2017.PubMed/NCBI
|
|
79
|
Li T, Chen Y, Zhang J and Liu S: LncRNA
TUG1 promotes cells proliferation and inhibits cells apoptosis
through regulating AURKA in epithelial ovarian cancer cells. Med
(Baltimore). 97:e121312018. View Article : Google Scholar
|
|
80
|
Yang LY, He CY, Chen XH, Su LP, Liu BY and
Zhang H: Aurora kinase A revives dormant laryngeal squamous cell
carcinoma cells via FAK/PI3K/Akt pathway activation. Oncotarget.
7:48346–48359. 2016.PubMed/NCBI
|
|
81
|
Schnepp RW, Khurana P, Attiyeh EF, Raman
P, Chodosh SE, Oldridge DA, Gagliardi ME, Conkrite KL, Asgharzadeh
S, Seeger RC, et al: A LIN28B-RAN-AURKA signaling network promotes
neuroblastoma tumorigenesis. Cancer Cell. 28:599–609. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang L, Zhao Z, Feng W, Ye Z, Dai W, Zhang
C, Peng J and Wu K: Long non-coding RNA TUG1 promotes colorectal
cancer metastasis via EMT pathway. Oncotarget. 7:51713–51719.
2016.PubMed/NCBI
|
|
83
|
Yin DD, Zhang EB, You LH, Wang N, Wang LT,
Jin FY, Zhu YN, Cao LH, Yuan QX, De W and Tang W: Downregulation of
lncRNA TUG1 affects apoptosis and insulin secretion in mouse
pancreatic β cells. Cell Physiol Biochem. 35:1892–1904. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yilmaz M and Christofori G: EMT, the
cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev.
28:15–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Vergara D, Merlot B, Lucot JP, Collinet P,
Vinatier D, Fournier I and Salzet M: Epithelial-mesenchymal
transition in ovarian cancer. Cancer Lett. 291:59–66. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Sun JF, Hu JY, Wang GJ, Yang Z, Zhao C,
Zhang X and Wang J: LncRNA TUG1 promoted KIAA1199 expression via
miR-600 to accelerate cell metastasis and epithelial-mesenchymal
transition in colorectal cancer. J Exp Clin Cancer Res. 37:1062018.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wang Y, Yang T, Zhang Z, Lu M, Zhao W,
Zeng X and Zhang W: Long non-coding RNA TUG1 promotes migration and
invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells.
Cancer Sci. 108:859–867. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ji TT, Huang X, Jin J, Pan SH and Zhuge
XJ: Inhibition of long non-coding RNA TUG1 on gastric cancer cell
transference and invasion through regulating and controlling the
expression of miR-144/c-Met axis. Asian Pac J Trop Med. 9:508–512.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
He C, Liu Z, Jin L, Zhang F, Peng X, Xiao
Y, Wang X, Lyu Q and Cai X: lncRNA TUG1-mediated Mir-142-3p
downregulation contributes to metastasis and the
Epithelial-to-Mesenchymal transition of hepatocellular carcinoma by
targeting ZEB1. Cell Physiol Biochem. 48:1928–1941. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Lin YH, Wu MH, Huang YH, Yeh CT, Cheng ML,
Chi HC, Tsai CY, Chung IH, Chen CY and Lin KH: Taurine up-regulated
gene 1 functions as a master regulator to coordinate glycolysis and
metastasis in hepatocellular carcinoma. Hepatology. 67:188–203.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Li C, Gao Y, Li Y and Ding D: TUG1
mediates methotrexate resistance in colorectal cancer via
miR-186/CPEB2 axis. Biochem Biophys Res Commun. 491:552–557. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Shafiee SM, Rasti M, Seghatoleslam A,
Azimi T and Owji AA: UBE2Q1 in a human breast carcinoma cell line:
Overexpression and interaction with p53. Asian Pac J Cancer Prev.
16:3723–3727. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Lee K, Jung ES, Choi YJ, Lee KY and Lee A:
Expression of pRb, p53, p16 and cyclin D1 and their clinical
implications in urothelial carcinoma. J Korean Med Sci.
25:1449–1455. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Jiang L, Zhang Q, Ren H, Ma S, Lu C, Liu
B, Liu J, Liang J, Li M and Zhu R: Dihydromyricetin enhances the
chemo-sensitivity of nedaplatin via regulation of the p53/Bcl-2
pathway in hepatocellular carcinoma cells. PLoS One.
10:e01249942015. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Elhag R, Mazzio EA and Soliman KF: The
effect of silibinin in enhancing toxicity of temozolomide and
etoposide in p53 and PTEN-mutated resistant glioma cell lines.
Anticancer Res. 35:1263–1269. 2015.PubMed/NCBI
|
|
96
|
Pilarsky C, Wenzig M, Specht T, Saeger HD
and Grützmann R: Identification and validation of commonly
overexpressed genes in solid tumors by comparison of microarray
data. Neoplasia. 6:744–750. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Okabe H, Satoh S, Kato T, Kitahara O,
Yanagawa R, Yamaoka Y, Tsunoda T, Furukawa Y and Nakamura Y:
Genome-wide analysis of gene expression in human hepatocellular
carcinomas using cDNA microarray: Identification of genes involved
in viral carcinogenesis and tumor progression. Cancer Res.
61:2129–2137. 2001.PubMed/NCBI
|
|
98
|
Li J, Zhang M, An G and Ma Q: Long
non-coding RNA TUG1 acts as a miR-26a sponge in human glioma cells.
Biochem Biophys Res Commun. 477:743–748. 2016. View Article : Google Scholar : PubMed/NCBI
|