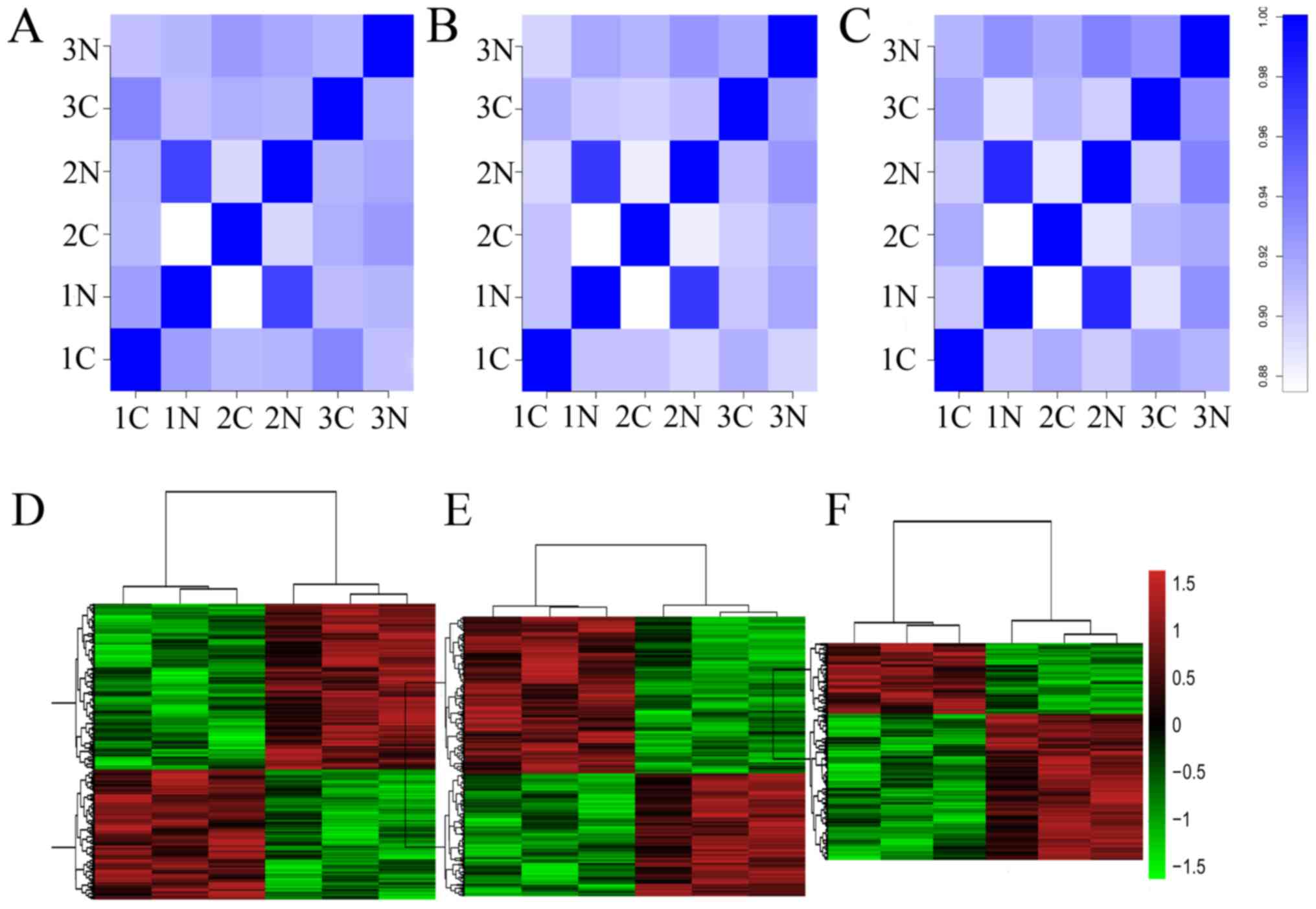
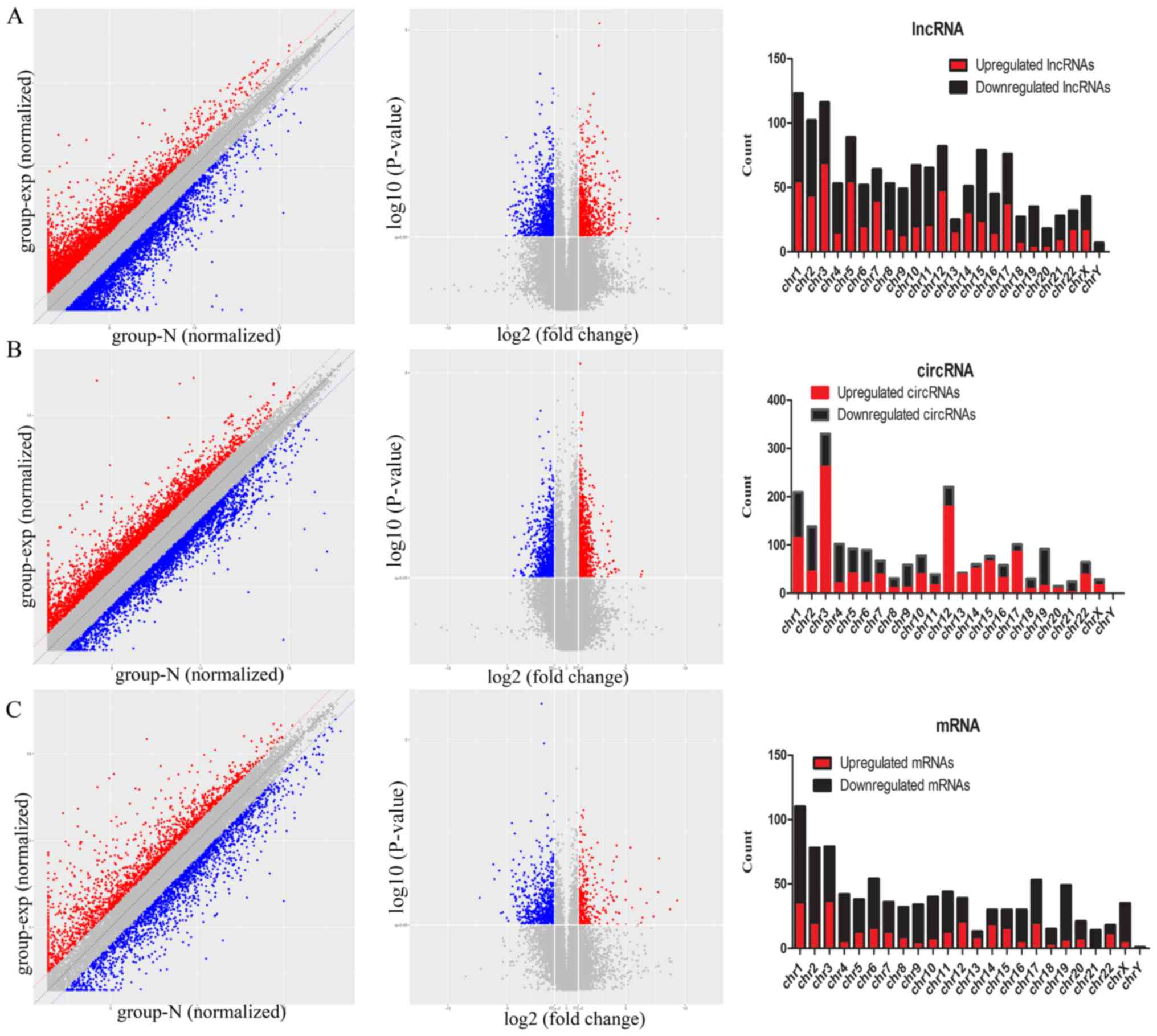
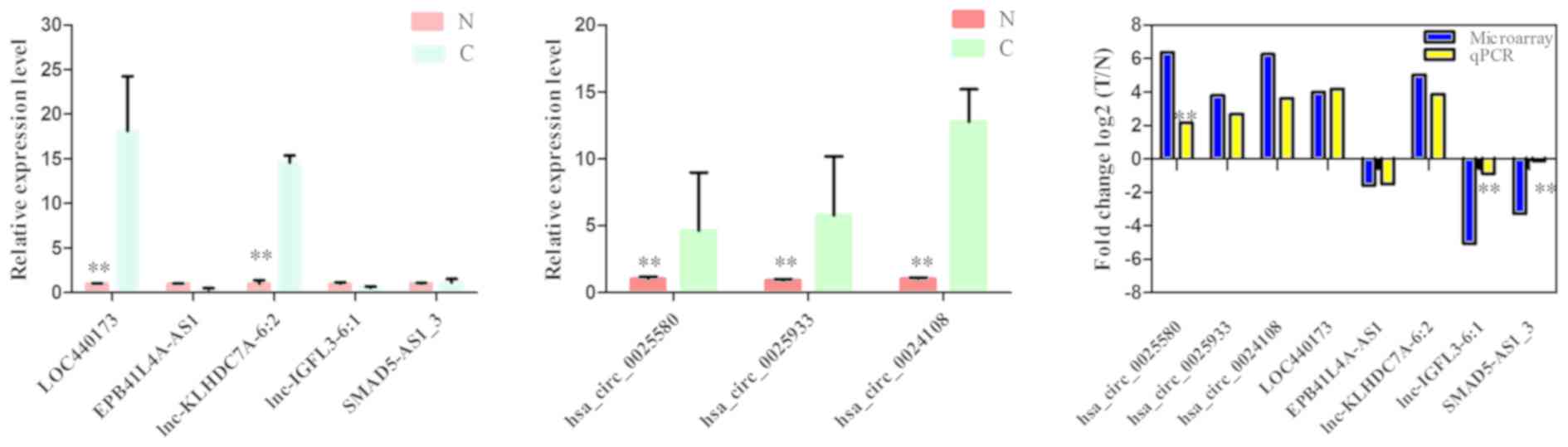
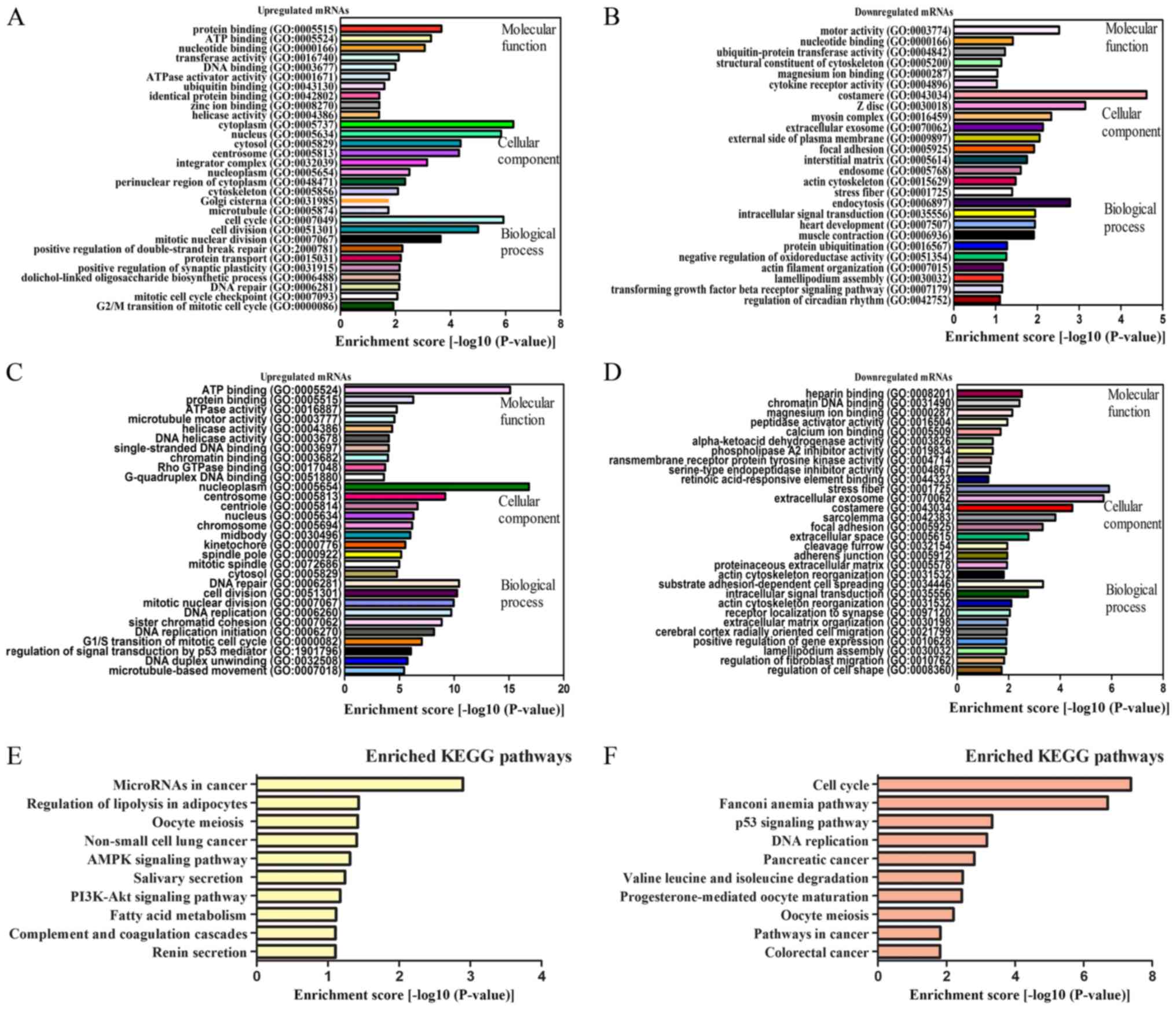
|
1
|
Forghanifard MM, Taleb Sh and Abbaszadegan
MR: Notch signaling target genes are directly correlated to
esophageal squamous cell carcinoma tumorigenesis. Pathol Oncol Res.
21:463–467. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zuo T, Zeng H, Zhang S
and He J: National cancer incidence and mortality in China 2012.
Chin J Cancer Res. 28:1–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Y: Epidemiology of esophageal
cancer. World J Gastroenterol. 19:5598–5606. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lander ES, Linton LM, Birren B, Nusbaum C,
Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rong D, Sun H, Li Z, Liu S, Dong C, Fu K,
Tang W and Cao H: An emerging function of circRNA-miRNAs-mRNA axis
in human diseases. Oncotarget. 8:73271–73281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Han X, Wang L, Ning Y, Li S and Wang Z:
Long non-coding RNA AFAP1-AS1 facilitates tumor growth and promotes
metastasis in colorectal cancer. Biol Res. 49:362016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou M, Hou Y, Yang G, Zhang H, Tu G, Du
YE, Wen S, Xu L, Tang X, Tang S, et al: LncRNA-Hh strengthen cancer
stem cells generationin Twist-positive breast cancer via activation
of Hedgehog signaling pathway. Stem Cells. 34:55–66. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu S, Wang P, You Z, Meng H, Mu G, Bai X,
Zhang G, Zhang J and Pang D: The longnon-coding RNAEPB41L4A-AS2
inhibits tumor proliferation and is associated with favorable
prognoses in breast cancer and other solid tumors. Oncotarget.
7:20704–20717. 2016.PubMed/NCBI
|
|
12
|
Lee S, Kopp F, Chang TC, Sataluri A, Chen
B, Sivakumar S, Yu H, Xie Y and Mendell JT: Noncoding RNA NORAD
regulates genomic stability by sequestering PUMILIO proteins. Cell.
164:69–80. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Geng Y, Jiang J and Wu C: Function and
clinical significance of circRNAs in solid tumors. J Hematol Oncol.
11:982018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jin X, Feng CY, Xiang Z, Chen YP and Li
YM: CircRNA expression pattern and circRNA-miRNA-mRNA network in
the pathogenesis of nonalcoholic steatohepatitis. Oncotarget.
7:66455–66467. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang M, Zhong Z, Lv M, Shu J, Tian Q and
Chen J: Comprehensive analysis of differentially expressed profiles
of lncRNAs and circRNAs with associated co-expression and ceRNA
networks in bladder carcinoma. Oncotarget. 7:47186–47200.
2016.PubMed/NCBI
|
|
19
|
Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z,
Yang J, Fan J, Liu L and Qin W: Hsa_circ_0001649: A circularRNA and
potential novel biomarker for hepatocellular carcinoma. Cancer
Biomark. 16:161–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peng L, Chen G, Zhu Z, Shen Z, Du C, Zang
R, Su Y, Xie H, Li H, Xu X, et al: CircularRNA ZNF609 functions as
a competitive endogenous RNA to regulate AKT3 expression by
sponging miR-150-5p in Hirschsprung's disease. Oncotarget.
8:808–818. 2017.PubMed/NCBI
|
|
21
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ and Liang L: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016.PubMed/NCBI
|
|
22
|
Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX,
Liao ZJ and Nan KJ: Over-expression of CircRNA_100876 in non-small
cell lung cancer and its prognostic value. Pathol Res Pract.
213:453–456. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y,
Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA circMTO1 acts as
the sponge of microRNA-9 to suppress hepatocellular carcinoma
progression. Hepatology. 66:1151–1164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen L, Zhang S, Wu J, Cui J, Zhong L,
Zeng L and Ge S: CircRNA_100290 plays a role in oral cancer by
functioning as a sponge of the miR-29 family. Oncogene.
36:4551–4561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolf PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou B and Yu JW: A novel identified
circular RNA, circRNA_010567, promotes myocardial fibrosis via
suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res
Commun. 487:769–775. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sobin LH, Gospodarowicz MK and Wittekind
C: Oesophagus including oesophagogastric junctionTNM classification
of malignant tumours. 7th. Oxford: Wiley-Blackwell; pp. 66–72.
2009
|
|
29
|
Coates DE, Zafar S and Milne TJ:
Quantitative real-time gene profiling of human alveolar
osteoblasts. Methods Mol Biol. 1537:447–459. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
The Gene Ontology Consortium, . Gene
ontology consortium: Going forward. Nucleic Acids Res. 43((Database
Issue)): D1049–D1056. 2015.PubMed/NCBI
|
|
31
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Petruk S, Sedkov Y, Riley KM, Hodgson J,
Schweisguth F, Hirose S, Jaynes JB, Brock HW and Mazo A:
Transcription of bxd noncoding RNAs promoted by trithorax represses
Ubx in cis by transcriptional interference. Cell. 127:1209–1221.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tafer H and Hofacker IL: RNAplex: A fast
tool for RNA-RNA interaction search. Bioinformatics. 24:2657–2663.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou M, Wang X, Li J, Hao D, Wang Z, Shi
H, Han L, Zhou H and Sun J: Prioritizing candidate disease-related
long non-coding RNAs by walking on the heterogeneous lncRNA and
disease network. Mol Biosyst. 11:760–7569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun J, Shi H, Wang Z, Zhang C, Liu L, Wang
L, He W, Hao D, Liu S and Zhou M: Inferring novel lncRNA-disease
associations based on a random walk model of a lncRNA functional
similarity network. Mol Biosyst. 10:2074–2081. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sahu A, Singhal U and Chinnaiyan AM: Long
noncoding RNAs in cancer: From function to translation. Trends
Cancer. 1:93–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li J, Yang J, Zhou P, Le Y, Zhou C, Wang
S, Xu D, Lin HK and Gong Z: Circular RNAs in cancer: Novel insights
into origins, properties, functions and implications. Am J Cancer
Res. 5:472–480. 2015.PubMed/NCBI
|
|
39
|
Wang X, Zhang Y, Huang L, Zhang J, Pan F,
Li B, Yan Y, Jia B, Liu H, Li S and Zheng W: Decreased expression
of hsa_circ_001988 in colorectal cancer and its clinical
significances. Int J Clin Exp Pathol. 8:16020–16025.
2015.PubMed/NCBI
|
|
40
|
Cai X, Hu X, Tan X, Cheng W, Wang Q, Chen
X, Guan Y, Chen C and Jing X: Metformin induced AMPK activation,
G0/G1 phase cell cycle arrest and the inhibition of growth of
esophageal squamous cell carcinomas in vitro and in vivo. PLoS One.
10:e01333492015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Peng HH, Zhang X and Cao PG: MMP-1/PAR-1
signal transduction axis and its prognostic impact in esophageal
squamous cell carcinoma. Braz J Med Biol Res. 45:86–92. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhou M, Diao Z, Yue X, Chen Y, Zhao H,
Cheng L and Sun J: Construction and analysis of dysregulated
lncRNA-associated ceRNA network identified novel lncRNA biomarkers
for early diagnosis of human pancreatic cancer. Oncotarget.
7:56383–56394. 2016.PubMed/NCBI
|
|
43
|
Zhu J, Chen X, Liao Z, He C and Hu X:
TGFBI protein high expression predicts poor prognosis in colorectal
cancer patients. Int J Clin Exp Pathol. 8:702–710. 2015.PubMed/NCBI
|