Introduction
Breast cancer is the most common malignant tumor and
a leading cause of cancer-related death among females worldwide.
Cases in China account for 12.2% of newly diagnosed breast cancers
and 9.6% of breast cancer-associated deaths worldwide in 2014
(1). Although advances have been
achieved in early diagnosis and systemic therapy, the prognosis of
patients with breast cancer remains poor. The identification of new
sensitive and specific biomarkers for the prognosis of patients
with breast cancer is therefore urgently needed.
Rho/Rac/cell division cycle 42-like (Rho-like)
GTPases are key regulators of multiple cell functions, including
cell polarity control, membrane transport, transcriptional
regulation, survival, adhesion and proliferation (2,3).
Rho-like GTPases are inactive in GDP-bound form and active in
GTP-bound form. GTPase activating proteins (RhoGAPs) are negative
regulators of Rho-like GTPases, which exert their functions by
catalyzing the conversion of the active GTP-bound state to the
inactive GDP-bound state. A family of genes encoding RhoGAPs
(ARHGAP) switch off Rho-like GTPases. Genetic alterations of
ARHGAP family genes are responsible for cancer biogenesis through
the dysregulation of Rho-like GTPases (2,3). Low
expression of ARHGAP7 is associated with poor prognosis in
patients with estrogen receptor (ER)-positive breast cancer with
further decrease in survival in patients with metastatic lesions
(4). ARHGAP15 is an
androgen-induced gene and has anti-tumor roles associated with the
Rac1 pathway (5). ARHGAP18
expression is associated with improved patient outcomes in invasive
breast cancer (6). Thus, researchers
and clinicians are increasingly considering ARHGAP
expression levels as a source of important clinical and predictive
therapeutic information.
Although previous studies have reported a general
expression profile of ARHGAP family genes in breast cancer,
several challenges remain in the identification of suitable and
novel biomarkers for precision treatment and prognosis. The present
study aimed to perform bioinformatics analysis of the
clinicopathological parameters and survival data associated with
ARHGAP family genes in patients with breast cancer by
pooling and analyzing several large online databases.
Materials and methods
Oncomine
Oncomine (http://www.oncomine.org) is an online database that
incorporates 715 datasets and 86,733 samples and aims to compute
gene expression signatures and extract biological insights from the
data for cancer research (7). All
the mentioned ARHGAP genes were queried in the database and
the results were filtered by selecting ‘breast cancer’ and ‘cancer’
vs. ‘normal’ analysis with the threshold of fold change ≥2,
P≤1×10−4, and gene rank ≥ top 10%.
Kaplan-Meier plotter
The Kaplan Meier Plotter (http://kmplot.com/analysis/) provides a powerful
platform for assessing the biological relationships between gene
expression levels and survival information including relapse-free
survival (RFS) and overall survival (OS) in patients with breast
cancer (8). P-values, hazard ratios
and 95% confidence intervals according to the mRNA expression level
(low or high) of each ARHGAP gene were obtained.
bcGenExMiner
The Breast Cancer Gene-Expression Miner v4.1
(bcGenExMiner v4.1; http://bcgenex.centregauducheau.fr/BC-GEM) is a mining
tool of published annotated genomics data (9,10). The
selected ARHGAP family genes were analyzed with clinical
parameters such as age, nodal status, the presence of estrogen
receptor (ER), progesterone receptor (PR) and
epidermal growth factor receptor-2 (HER-2),
Scarff-Bloom-Richardson (SBR) grade and Nottingham prognostic index
(NPI). Prognostic values of metastatic relapse event and
ARHGAP genes were calculated using the prognostic module
(9,10).
cBioPortal
The cBioPortal (http://www.cbioportal.org) database offers
visualization, analysis and download of large-scale cancer genomics
datasets (11,12). To analyze the ARHGAP-centered
regulation system, a network of the ARHGAP family and the
neighboring genes was generated in cBioPortal.
Statistical analysis
According to protocols of the aforementioned tools,
mRNA levels of ARHGAP in breast cancer and normal tissues in each
individual dataset were analyzed using Student's t-test.
Kaplan-Meier survival analysis was performed to compare patient
survival based on ARHGAP expression levels by log-rank test.
Global significant difference between groups of clinical parameters
was assessed by Welch's and Dunnett-Tukey-Kramer's tests. Data are
presented as the mean ± standard error of mean (SEM). P<0.05 was
considered to indicate a statistically significant difference.
Results
Dysregulated expression of ARHGAP
genes in patients with breast cancer
The expression of ARHGAP family genes in were
evaluated in 20 common types of cancer, and their levels were
compared to normal individuals using the Oncomine database. Lower
expression levels (blue) of ARHGAP6, 7, 10, 14, 19, 23 and
24 and higher expression levels (red) of ARHGAP9, 11, 15,
18 and 30 were observed in breast cancer samples
compared with normal tissues. ARHGAP4, 8, 25 and 29
were neither upregulated nor downregulated in patients with breast
cancer compared with healthy individuals (Fig. 1).
Dysregulated ARHGAP genes in RFS and
OS of patients with breast cancer
The survival data of ARHGAP family genes were
analyzed using the Kaplan-Meier Plotter. The Kaplan-Meier curves
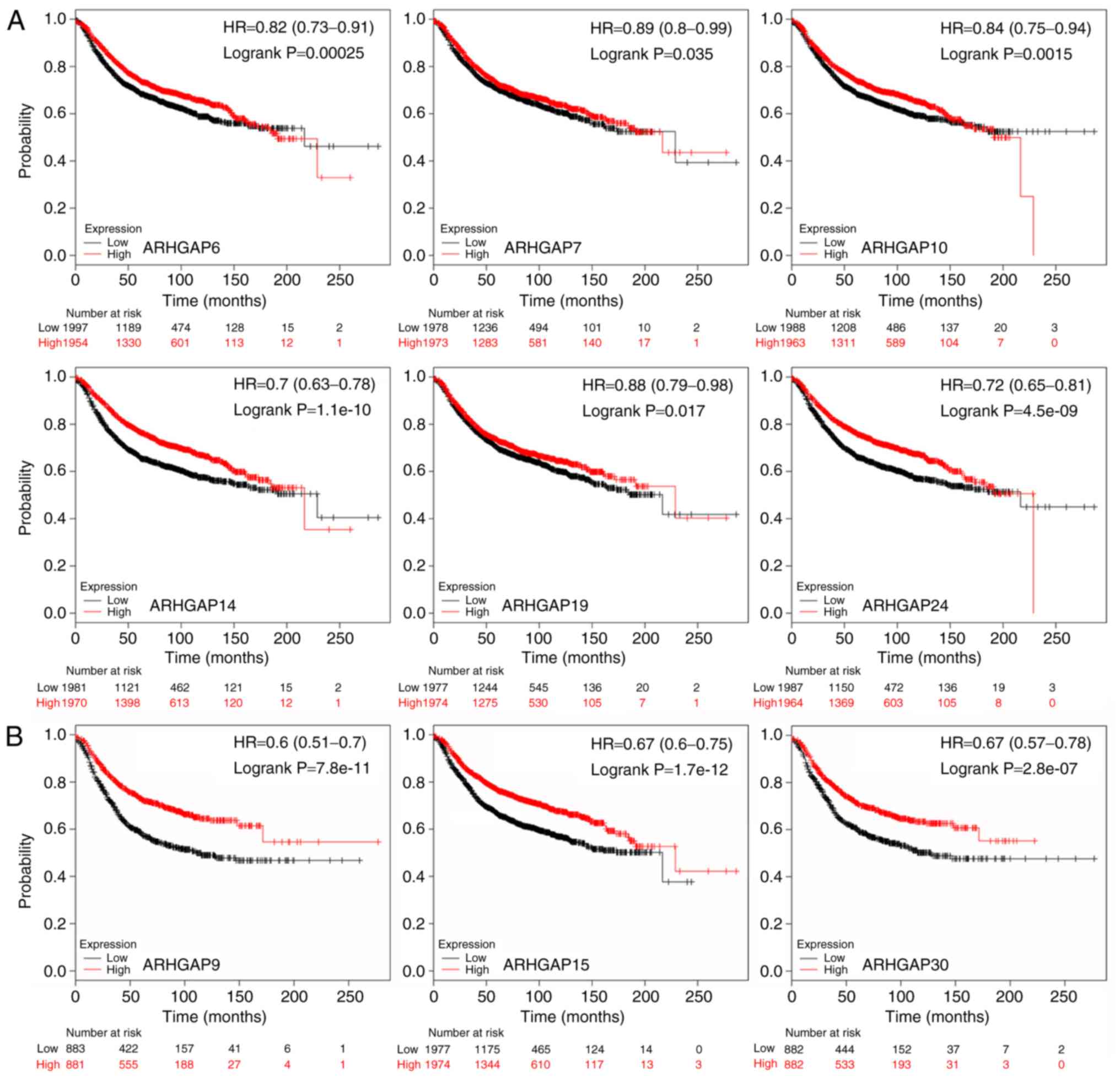
demonstrated that reduced ARHGAP6, 7, 10, 14, 19 and
24 mRNA levels were significantly associated with poor RFS
(Table I; Fig. 2A). Patients with high expression
levels of ARHGAP9, 15 and 30 exhibited favorable RFS
(Table I; Fig. 2B). In addition, low expression of
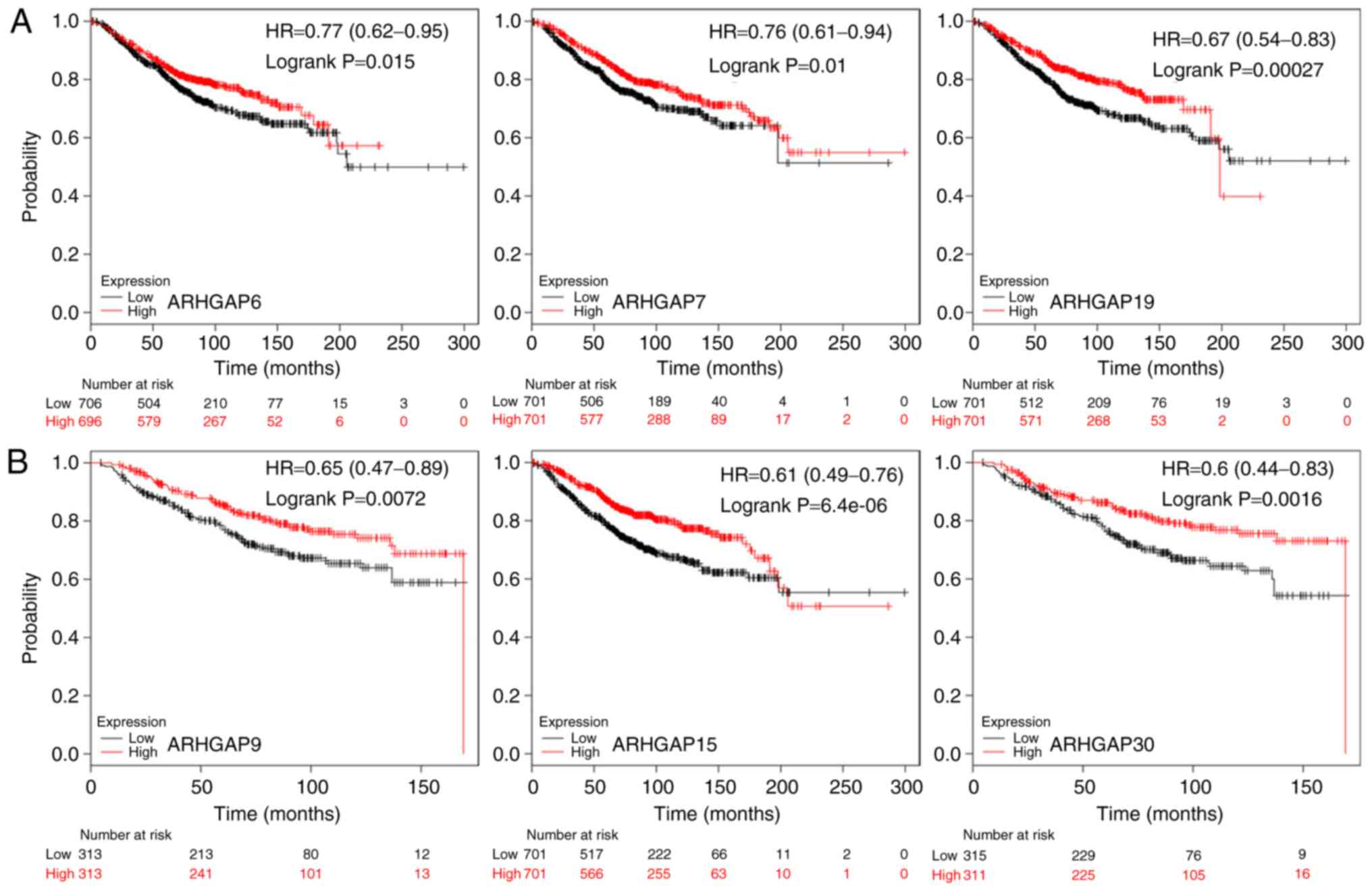
ARHGAP6, 7 and 19 was associated with poor OS
(Table II; Fig. 3A), whereas high expression of
ARHGAP9, 15 and 30 were associated with preferable OS
(Table II; Fig. 3B). To further verify the role of
ARHGAP family genes in breast cancer prognosis, the
bc-GenExMiner online software was used; ARHGAP15 exhibited
the most significant positive effect on patient metastatic
relapse-free survival, and the expression levels of ARHGAP9,
19 and 30 were associated with improved metastatic
relapse-free survival compared with patients in the respective low
expression groups (Table III).
 | Table I.Prognostic association between
ARHGAP family gene expression in breast cancer and
relapse-free survival based on Kaplan-Meier Plotter analysis. |
Table I.
Prognostic association between
ARHGAP family gene expression in breast cancer and
relapse-free survival based on Kaplan-Meier Plotter analysis.
| Gene name | Cut-off value | Expression (range
of probe) | P-value | HR (95% CI) | N |
|---|
| ARHGAP4 | 125 | 6-1003 | >0.0001 | 0.75
(0.67–0.83) | 3,951 |
| ARHGAP6 | 136 | 3-2602 | 0.0003 | 0.82
(0.73–0.91) | 3,951 |
| ARHGAP7 | 901 | 26-10898 | 0.0350 | 0.89
(0.80–0.99) | 3,951 |
| ARHGAP8 | 867 | 6-8528 | 0.0004 | 0.82
(0.74–0.91) | 3,951 |
| ARHGAP9 | 398 | 24-4139 | >0.0001 | 0.60
(0.51–0.70) | 1,764 |
|
ARHGAP10 | 271 | 16-1124 | 0.0015 | 0.84
(0.75–0.94) | 3,951 |
|
ARHGAP11A | 45 | 1-806 | 0.1300 | 1.09
(0.97–1.21) | 3,951 |
|
ARHGAP14 | 221 | 6-1787 | >0.0001 | 0.70
(0.63–0.78) | 3,951 |
|
ARHGAP15 | 316 | 4-4586 | >0.0001 | 0.67
(0.60–0.75) | 3,951 |
|
ARHGAP18 | 157 | 3-1189 | 0.0010 | 0.77
(0.66–0.90) | 1,764 |
|
ARHGAP19 | 246 | 12-1183 | 0.0170 | 0.88
(0.79–0.98) | 3,951 |
|
ARHGAP23 | 237 | 9-5617 | 0.0070 | 0.81
(0.69–0.94) | 1,764 |
|
ARHGAP24 | 110 | 3-1758 | >0.0001 | 0.72
(0.65–0.81) | 3,951 |
|
ARHGAP25 | 171 | 3-4021 | 0.0051 | 0.86
(0.77–0.95) | 3,951 |
|
ARHGAP29 | 112 | 1-1992 | >0.0001 | 0.57
(0.49–0.67) | 1,764 |
|
ARHGAP30 | 441 | 21-3316 | >0.0001 | 0.67
(0.57–0.78) | 1,764 |
 | Table II.Prognostic association between
ARHGAP family gene expression in breast cancer and overall
survival based on Kaplan-Meier Plotter analysis. |
Table II.
Prognostic association between
ARHGAP family gene expression in breast cancer and overall
survival based on Kaplan-Meier Plotter analysis.
| Gene name | Cut-off value | Expression (range
of probe) | P-value | HR (95% CI) | N |
|---|
| ARHGAP4 | 120 | 10-699 | 0.0670 | 0.82
(0.66–1.01) | 1,402 |
| ARHGAP6 | 157 | 3-3919 | 0.0150 | 0.77
(0.62–0.95) | 1,402 |
| ARHGAP7 | 1,031 | 37-10272 | 0.0100 | 0.76
(0.61–0.94) | 1,402 |
| ARHGAP8 | 850 | 7-8528 | 0.6000 | 1.06
(0.85–1.31) | 1,402 |
| ARHGAP9 | 414 | 41-3927 | 0.0072 | 0.65
(0.47–0.89) | 626 |
|
ARHGAP10 | 298 | 21-1124 | 0.6300 | 1.05
(0.85–1.31) | 1,402 |
|
ARHGAP11A | 46 | 1-806 | 0.7200 | 1.04
(0.84–1.29) | 1,402 |
|
ARHGAP14 | 216 | 6-1540 | 0.0550 | 0.81
(0.65–1.00) | 1,402 |
|
ARHGAP15 | 363 | 4-4586 | >0.0001 | 0.61
(0.49–0.76) | 1,402 |
|
ARHGAP18 | 170 | 5-1189 | 0.0660 | 0.75
(0.55–1.02) | 626 |
|
ARHGAP19 | 235 | 16-1078 | 0.0003 | 0.67
(0.54–0.83) | 1,402 |
|
ARHGAP23 | 219 | 9-4750 | 0.1300 | 1.27
(0.93–1.74) | 626 |
|
ARHGAP24 | 115 | 5-1758 | 0.4500 | 0.92
(0.74–1.14) | 1,402 |
|
ARHGAP25 | 172 | 7-2335 | 0.0006 | 0.68
(0.55–0.85) | 1,402 |
|
ARHGAP29 | 126 | 2-1388 | 0.0250 | 0.70
(0.51–0.96) | 626 |
|
ARHGAP30 | 470 | 21-2542 | 0.0016 | 0.60
(0.44–0.83) | 626 |
 | Table III.Prognostic association ARHGAP
family gene expression in breast cancer between and metastatic
relapse based on bc-GenExMiner analysis. |
Table III.
Prognostic association ARHGAP
family gene expression in breast cancer between and metastatic
relapse based on bc-GenExMiner analysis.
| Gene name | P-value | HR | 95% CI | N | Metastatic
relapse |
|---|
| ARHGAP4 | 0.4483 | 0.98 | 0.92–1.04 | 3,825 | 993 |
| ARHGAP6 | 0.4608 | 0.97 | 0.89–1.06 | 3,500 | 907 |
| ARHGAP7 | 0.1422 | 0.94 | 0.87–1.02 | 3,924 | 1,023 |
| ARHGAP8 | 0.1410 | 1.09 | 0.97–1.21 | 1,345 | 340 |
| ARHGAP9 | 0.0244 | 0.89 | 0.81–0.99 | 1,721 | 438 |
|
ARHGAP10 | 0.3605 | 1.03 | 0.96–1.11 | 3,456 | 878 |
|
ARHGAP11A | 0.0099 | 1.09 | 1.02–1.17 | 3,826 | 993 |
|
ARHGAP14 | <0.0001 | 1.20 | 1.11–1.29 | 3,610 | 911 |
|
ARHGAP15 | <0.0001 | 0.84 | 0.79–0.89 | 3,701 | 966 |
|
ARHGAP18 | <0.0001 | 0.82 | 0.75–0.90 | 2,016 | 539 |
|
ARHGAP19 | 0.0006 | 0.89 | 0.84–0.95 | 3,923 | 1,023 |
|
ARHGAP23 | 0.0574 | 1.15 | 1.00–1.33 | 1,425 | 358 |
|
ARHGAP24 | 0.1126 | 0.94 | 0.88–1.01 | 3,845 | 1,006 |
|
ARHGAP25 | 0.0009 | 0.89 | 0.83–0.95 | 3,826 | 993 |
|
ARHGAP29 | 0.0649 | 0.94 | 0.88–1.00 | 3,925 | 1,023 |
|
ARHGAP30 | 0.0490 | 0.90 | 0.81–1.00 | 1,862 | 491 |
ARHGAP genes and clinicopathological
characteristics of patients with breast cancer
By comparing the aforementioned databases, the
expression levels of the overlapped genes ARHGAP9, 15, 19
and 30 were analyzed between different patient groups based
on clinicopathological characteristics. The SBR grade, which
evaluates tubule formation, nuclear characteristics of pleomorphism
and mitotic index, is an important prognostic factor in breast
cancer (13). Patients with high
grade (SBR3) tumors tended to express high levels of ARHGAP9
and 30 and low levels of ARHGAP19 than lower grade
(SBR1) tumors (Fig. 4A). The NPI is
based on histopathological factors and is used to stratify patients
with breast cancer into prognostic groups (14); low expression of ARHGAP19 was
associated with NPI (Fig. 4B). No
significant differences were observed between the ≤51 and >51
years groups, with an exception for ARHGAP19, which was
expressed at low levels in the >51 years group. Patients with
ER-positive or PR-positive breast cancer exhibited lower expression
levels of ARHGAP9, 15 and 30 compared with patients
with ER-negative or PR-negative status. Patients with
HER-2-positive status exhibited higher expression levels of
ARHGAP9 and 30 compared with patients with
HER-2-negative status. In addition, ARHGAP9, 15, 19 and
30 expression levels were significantly elevated in patients
with triple-negative breast cancer compared with patients without
triple-negative breast cancer. No significant association was
observed between nodal status and ARHGAP9, 15, 19 and
30 expression levels (Fig.
5).
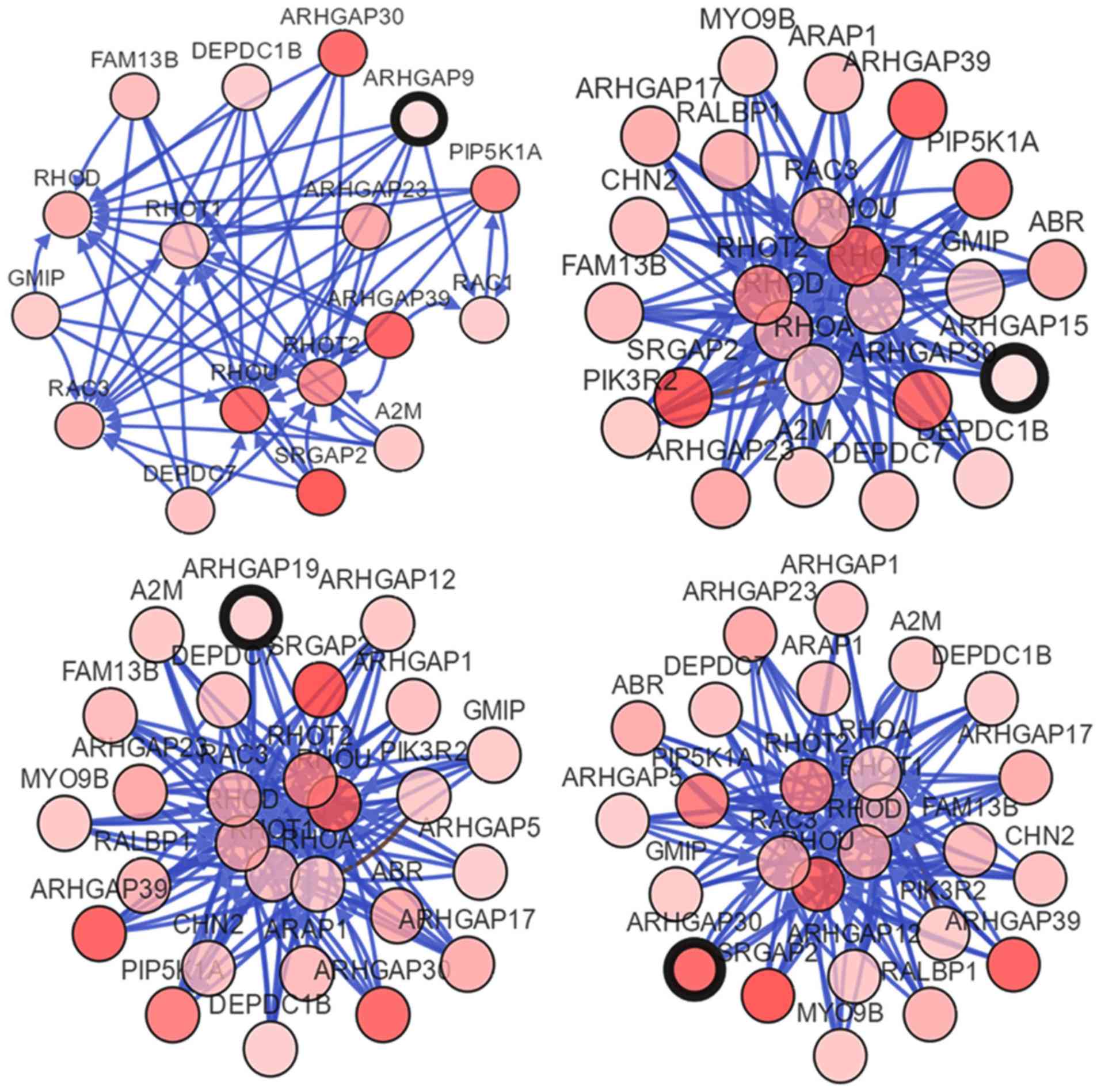
Construction of the ARHGAP gene
network
To better visualize the potential genes interacting
with ARHGAP9, 15, 19 and 30, a gene network was
constructed using the cBioPortal online database. ARHGAP23
and 39 were connected to ARHGAP9, 15, 19 and
30. Of note, ARHGAP9, 15 and 19 also
interacted with ARHGAP30 (Fig.
6).
Discussion
Rho-like GTPases are involved in various cell
functions and are negative regulators of Rho proteins; RhoGAPs
serve significant roles in multiple aspects of tumor biology
including gene expression, cell cycle, survival, migration and
invasion (3,6). ARHGAP, a group of family genes
encoding RhoGAPs that switch off Rho-like GTPases, have been
extensively studied since the discovery that genetic alterations of
ARHGAP family genes are responsible for breast cancer
biogenesis (2,3). However, several challenges remain
regarding the identification of suitable and novel biomarkers for
precision treatment and prognosis. To best of our knowledge, this
is the first report to characterize specific ARHGAP genes
with prognostic value and biological function in breast cancer
using bioinformatics analysis.
Although the involvement of ARHGAP in cancer
progression is becoming increasingly apparent, the association
between ARHGAP expression and most cancer types has not been
fully characterized. Therefore, a thorough study to determine the
expression of ARHGAP in different types of cancer is needed.
Researchers and clinicians are increasingly regarding ARHGAP
family genes as important clinical and predictive therapeutic
information. In the present study, a total of 16 ARHGAP family
genes were evaluated in breast cancer samples and compared with
those in normal tissues using the Oncomine database; the results
demonstrated that ARHGAP6, 7, 10, 14, 19, 23 and 24
were downregulated, ARHGAP9, 11, 15, 18 and 30 were
upregulated, and ARHGAP4, 8, 25 and 29 exhibited no
dysregulation. The number of significant unique analyses was small
for ARHGAP6, 9, 10, 14, 15, 23 and 30 or even zero in
ARHGAP4, 8, 25 and 29; however, bioinformatics
analysis was performed using all genes to identify several suitable
and novel biomarkers among ARGHAP family genes. The results
of the Kaplan-Meier survival analysis demonstrated that reduced
ARHGAP6, 7 and 19 were associated with poor RFS and
OS, whereas increased ARHGAP9, 15 and 30 were
associated with preferable RFS and OS. In addition, bc-GenExMiner
online software provided metastatic relapse data, which revealed
thatARHGAP9, 15, 18, 19, 25 and 30 were associated
with favorable prognosis, whereas high expression levels of
ARHGAP11A and 14 exerted negative effects on patient
prognosis. Therefore, ARHGAP9, 15, 19 and 30 were
identified as potential prognostic targets for breast cancer by
comparing databases and the overlapped genes.
ARHGAP9, which is a mitogen-activated protein
kinase-docking protein, inhibits mitogen-activated protein kinase 1
and p38α activation through WW domain binding (15,16).
ARHGAP9 has been demonstrated to suppress the migration and
invasion of hepatocellular carcinoma cells by upregulating forkhead
box J2 and E-cadherin (17). In
addition, downregulated ARHGAP9 is associated with breast
cancer risk and suppresses the proliferation, migration and
invasion of breast cancer cells (18). The present study also demonstrated
that high levels of ARHGAP9 were associated with RFS and OS
advantages in patients with breast cancer patients and may be a
promising prognostic factor.
ARHGAP15, a Rac-specific RhoGAP described in
2003, serves a dual role in inhibiting small GTPase signaling
(19,20). A previous study has demonstrated that
decreased expression of ARHGAP15 promotes the development of
colorectal cancer through the PTEN/AKT/forkhead box protein O1 axis
(21). In addition, ARHGAP15
is an androgen-induced gene and serves an antitumor function
associated with the Rac1 pathway (5). These results, along with the results of
the survival analysis based on ARHGAP15 expression levels in
the present study, suggested that ARHGAP15 may serve as a
tumor suppressor during breast cancer progression and
metastasis.
ARHGAP30, a Rac1- and RhoA-specific RhoGAP,
is a Wrch-1-interacting protein involved in actin dynamics and cell
adhesion (22). ARHGAP30 is
required for p53 acetylation and functional activation in
colorectal cancer; ectopic expression of ARHGAP30 induced
p53 activation and efficiently suppressed tumor growth in an in
vivo xenograft study (23).
Therefore, ARHGAP30 may be a prognostic marker and a
potential therapeutic target for cancer, which is consistent with
the results of the bioinformatics analysis in the present
study.
ARHGAP19 is predominantly expressed in
hematopoietic cells and controls cytokinesis and chromosome
segregation in T lymphocytes (24).
In the present study, low ARHGAP19 expression levels were
associated with poor RFS and OS. In addition, bc-GenExMiner
software provided metastatic relapse data, which demonstrated that
high ARHGAP19 expression was associated with favorable
metastatic relapse-free survival. These results, along with the
observation of decreased ARHGAP19 expression in patients
with high-grade tumors compared with patients with low-grade
tumors, opposed the result that ARHGAP19 expression levels
were elevated in patients with triple-negative breast cancer. Since
there is limited information on ARHGAP19 expression in
breast cancer, further studies are necessary to determine how
ARHGAP19 is involved in breast cancer biology and
progression.
Patients with ER- or PR-positive breast cancer are
often treated with drugs that block estrogen effects and generally
exhibit good prognosis with compared with ER- or PR-negative
patients. The results of the present study demonstrated that
ARHGAP9, 15 and 30 exhibited lower expression in ER-
or PR-positive cancer compared with ER- or PR-negative cases, which
contradicted the earlier conclusion that ARHGAP9, 15 and
30 acts as tumor suppressors. This difference may be due to
the ARHGAP expression profiles in the dataset being
primarily from RNA sequences, as well as due to differences in the
clinical samples and experimental conditions. Further
investigations are required to precisely elucidate the
physiological relevance of ARHGAP9, 15, 19 and
30.
In conclusion, the results of the present study
suggested that ARHGAP9, 15, 19 and 30, compared with
other ARHGAP family genes, might be promising targets with
prognostic value and biological function for precision treatment in
patients with breast cancer. Further experiments and clinical
trials are required to validate the value of these genes.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of China (grant no. 81702591) and the Natural
Science Foundation of Jiangsu Province (grant no. BK20170294), and
the Post-doctoral Foundation of Jiangsu Province (grant no.
2019K161).
Availability of data and materials
The datasets analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
WXC, ML, LC, YLZ and HD conceived and designed the
experiments. WXC, LC and QQ performed the experiments. QQ, LYX and
LS analyzed the data. YLZ and HD contributed to reagents, materials
and analysis tools. WXC and ML wrote the manuscript. HD gave final
approval of the version to be published.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast
cancer in China. Lancet Oncol. 15:e279–e289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vega FM and Ridley AJ: Rho GTPases in
cancer cell biology. FEBS Lett. 582:2093–2101. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Etienne-Manneville S and Hall A: Rho
GTPases in cell biology. Nature. 420:629–635. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gökmen-Polar Y, True JD, Vieth E, Gu Y, Gu
X, Qi GD, Mosley AL and Badve SS: Quantitative phosphoproteomic
analysis identifies novel functional pathways of tumor suppressor
DLC1 in estrogen receptor positive breast cancer. PLoS One.
13:e02046582018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takagi K, Miki Y, Onodera Y, Ishida T,
Watanabe M, Sasano H and Suzuki T: ARHGAP15 in human breast
carcinoma: A potent tumor suppressor regulated by androgens. Int J
Mol Sci. 19:E8042018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Aleskandarany MA, Sonbul S, Surridge R,
Mukherjee A, Caldas C, Diez-Rodriguez M, Ashankyty I, Albrahim KI,
Elmouna AM, Aneja R, et al: Rho-GTPase activating-protein 18: A
biomarker associated with good prognosis in invasive breast cancer.
Br J Cancer. 117:1176–1184. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jézéquel P, Frénel JS, Campion L,
Guérin-Charbonnel C, Gouraud W, Ricolleau G and Campone M:
bc-GenExMiner 3.0: New mining module computes breast cancer gene
expression correlation analyses. Database (Oxford).
2013:bas0602013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jézéquel P, Campone M, Gouraud W,
Guérin-Charbonnel C, Leux C, Ricolleau G and Campion L:
bc-GenExMiner: An easy-to-use online platform for gene prognostic
analyses in breast cancer. Breast Cancer Res Treat. 131:765–775.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Le Doussal V, Tubiana-Hulin M, Friedman S,
Hacene K, Spyratos F and Brunet M: Prognostic value of histologic
grade nuclear components of Scarff-Bloom-Richardson (SBR). An
improved score modification based on a multivariate analysis of
1262 invasive ductal breast carcinomas. Cancer. 64:1914–1921. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Haybittle JL, Blamey RW, Elston CW,
Johnson J, Doyle PJ, Campbell FC, Nicholson RI and Griffiths K: A
prognostic index in primary breast cancer. Br J Cancer. 45:361–366.
1982. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ang BK, Lim CY, Koh SS, Sivakumar N, Taib
S, Lim KB, Ahmed S, Rajagopal G and Ong SH: ArhGAP9, a novel MAP
kinase docking protein, inhibits Erk and p38 activation through WW
domain binding. J Mol Signal. 2:12007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Furukawa Y, Kawasoe T, Daigo Y, Nishiwaki
T, Ishiguro H, Takahashi M, Kitayama J and Nakamura Y: Isolation of
a novel human gene, ARHGAP9, encoding a rho-GTPase activating
protein. Biochem Biophys Res Commun. 284:643–649. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang H, Tang QF, Sun MY, Zhang CY, Zhu
JY, Shen YL, Zhao B, Shao ZY, Zhang LJ and Zhang H: ARHGAP9
suppresses the migration and invasion of hepatocellular carcinoma
cells through up-regulating FOXJ2/E-cadherin. Cell Death Dis.
9:9162018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang T and Ha M: Silencing ARHGAP9
correlates with the risk of breast cancer and inhibits the
proliferation, migration, and invasion of breast cancer. J Cell
Biochem. 119:7747–7756. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Radu M, Rawat SJ, Beeser A, Iliuk A, Tao
WA and Chernoff J: ArhGAP15, a Rac-specific GTPase-activating
protein, plays a dual role in inhibiting small GTPase signaling. J
Biol Chem. 288:21117–21125. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Seoh ML, Ng CH, Yong J, Lim L and Leung T:
ArhGAP15, a novel human RacGAP protein with GTPase binding
property. FEBS Lett. 539:131–137. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pan S, Deng Y, Fu J, Zhang Y, Zhang Z, Ru
X and Qin X: Decreased expression of ARHGAP15 promotes the
development of colorectal cancer through PTEN/AKT/FOXO1 axis. Cell
Death Dis. 9:6732018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Naji L, Pacholsky D and Aspenström P:
ARHGAP30 is a Wrch-1-interacting protein involved in actin dynamics
and cell adhesion. Biochem Biophys Res Commun. 409:96–102. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang J, Qian J, Hu Y, Kong X, Chen H, Shi
Q, Jiang L, Wu C, Zou W, Chen Y, et al: ArhGAP30 promotes p53
acetylation and function in colorectal cancer. Nat Commun.
5:47352014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
David MD, Petit D and Bertoglio J: The
RhoGAP ARHGAP19 controls cytokinesis and chromosome segregation in
T lymphocytes. J Cell Sci. 127:400–410. 2014. View Article : Google Scholar : PubMed/NCBI
|